In vitro diagnostic kit for identification of human papillomavirus in clinical samples
A technology of human papillomavirus and samples, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, etc., and can solve the problems of expensive equipment and laborious operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1: Preparation of 'lattice-tube'
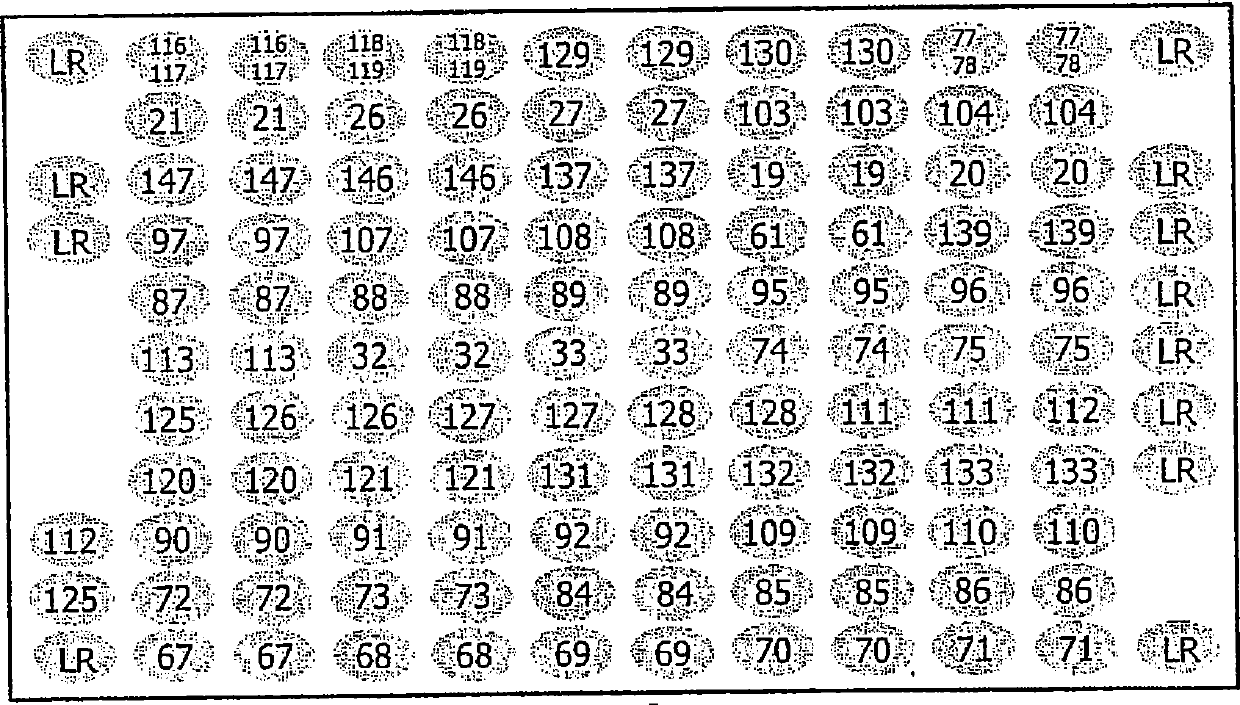
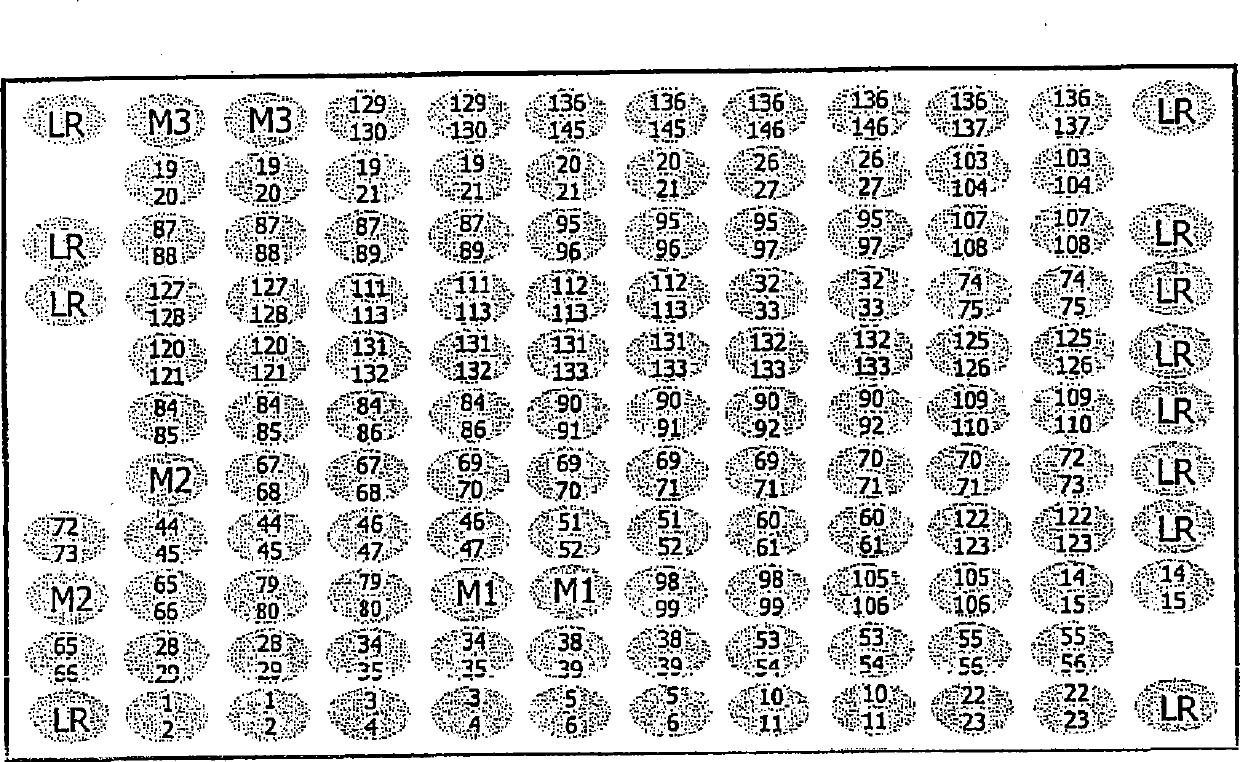
[0063] The 'array tube' of the present invention was fabricated at CLONDIAG Chip Technology GmbH (Jena, Germany) as follows. Standard reaction test tubes from Eppendorf made of polypropylene and having a specified receiving volume of 1.5 ml were modified by remelting, so that in the tubes molded with adhesive edges were used for microarray supports. Open recess.
[0064] Microarrays to be inserted into these tubes were produced by using a MicroGrid II Arrayer (BioRobotics, Cambridge, UK). A probe consisting of a 5' terminal amino-modified oligonucleotide having a sequence from the Sequence Listing was arranged at a defined site on the epoxy glass surface of a glass slide (slide size: 75mm x 25mm), and covalently fixed. A single microarray contains 12 x 10 = 120, or 12 x 11 = 132 specific positions where oligonucleotides can be placed. The positions are spaced 0.2mm apart so that the DNA libraries contained in each microarra...
Embodiment 2
[0067] Example 2: Preparation of DNA samples
[0068] 2.1. HPV DNA standard
[0069] HPV DNAs used to assess the specificity and sensitivity of type-specific probes were those containing amplified L1 regions (HPV types 6, 11, 13, 16, 18, 26, 31, 33, 35, 39, 40, 42, 44, 45, 51, 52, 53, 54, 56, 58, 61, 62, 66, 68, 70, 71, 72, 73, 81, 82, 83, 84, 85 and 89) recombinant plasmids, Or DNAs extracted from clinical samples whose amplified L1 region was further characterized by DNA sequencing. Recombinant plasmids were constructed by molecular cloning techniques. Briefly, the L1 region amplified from each HPV type was cloned into Easy carrier. Purified preparations obtained from each recombinant plasmid were further characterized by sequence analysis. 1-10 pg of plasmid DNA was used for the evaluation of specificity experiments.
[0070] DNA from the K562 cell line (Catalog No. DD2011, Promega Corporation, Madison, WI, USA) was used to evaluate the specificity and sensitivity of...
Embodiment 3
[0078] Embodiment 3: PCR amplification
[0079] PCR amplification using consensus primers MY11 and MY09 was performed (Manos et al., Molecular Diagnostics of Human Cancer; Furth M, Greaves MF, eds.; Cold Spring Harbor Press. 1989, Vol. 7:209-214). Also included in the PCR reaction is a third primer, HMB01, which is typically used in combination with MY09 and MY11 to amplify HPV type 51 that cannot be amplified efficiently using only MY09 and MY11 (Hildesheim et al., J Infect Dis. Journal) 1994, 169; 235-240). Briefly, PCR amplification was performed in 50 μl final volume reactions containing 10 mM Tris-HCl pH 8.3, 50 mM KCl, 1 mM MgCl 2 , 0.3 μM each of primers MY09 and MY11 (SEQ ID NO142 and 143), 0.03 μM of primer HMB01 (SEQ ID NO 144), 200 μM of each dNTP, 4 units of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA, USA), and 5 μl of each HPV DNA standard from Example 2.1. or clinical sample DNA from Example 2.2. To test the suitability of sample DNA, 0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com