Microorganism for producing beta-1,3-glucanase and application thereof
A glucanase, microorganism technology, applied in the direction of hydrolase, bacteria, etc., can solve the problem of insufficient bacteria source
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] The isolation and screening of chitosanase producing bacteria H12, the steps are as follows:
[0039] The soil samples were collected in Baotu Spring Brewery, planted on the screening medium, and cultivated at 30°C for 3 days, picked the edge of the colony with a large transparent circle of the degraded yeast cell wall, and transferred it to the screening medium again, at 30°C Cultivate for 3 days, pick out the thalli at the edge of the transparent circle of the degraded yeast cell wall, and separate them by streaking to obtain pure colony strains. The strain was identified as chitosanase-producing strain H12 (Mitsuaria chitosanitabida H12), and its 16SrRNA gene sequence is shown in Seq. Nol.
[0040] The screening medium used is: Angel yeast 5.0g, CaCl 2 2H 2 O 1.36g, agar 10g, add water to 1L, sterilize at 121°C for 30min.
Embodiment 2
[0041] Embodiment 2: Fermentation culture and β-1,3-glucanase activity determination of chitosanase producing bacteria H12
[0042] The fermentation medium is: 1-2% (w / v) Angel yeast, pH 6-7, autoclaved at 121° C. for 30 minutes. Inoculate the chitosanase-producing bacteria H12 obtained by the separation and screening of the present invention at an inoculation amount of 3-5% (v / v), culture at 30° C. at 160 rpm for 15h-18h on a shaking table to obtain crude enzyme liquid.
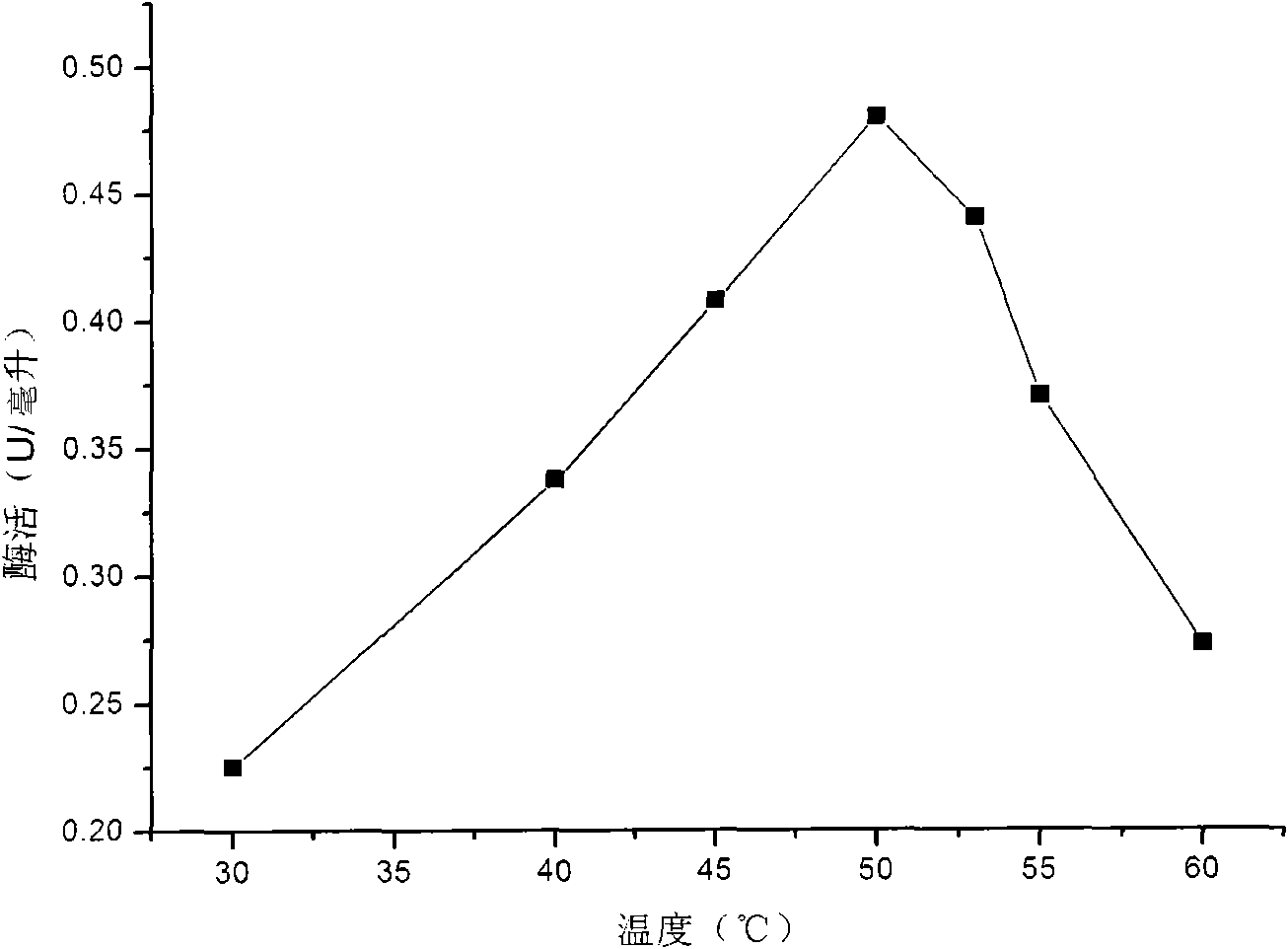
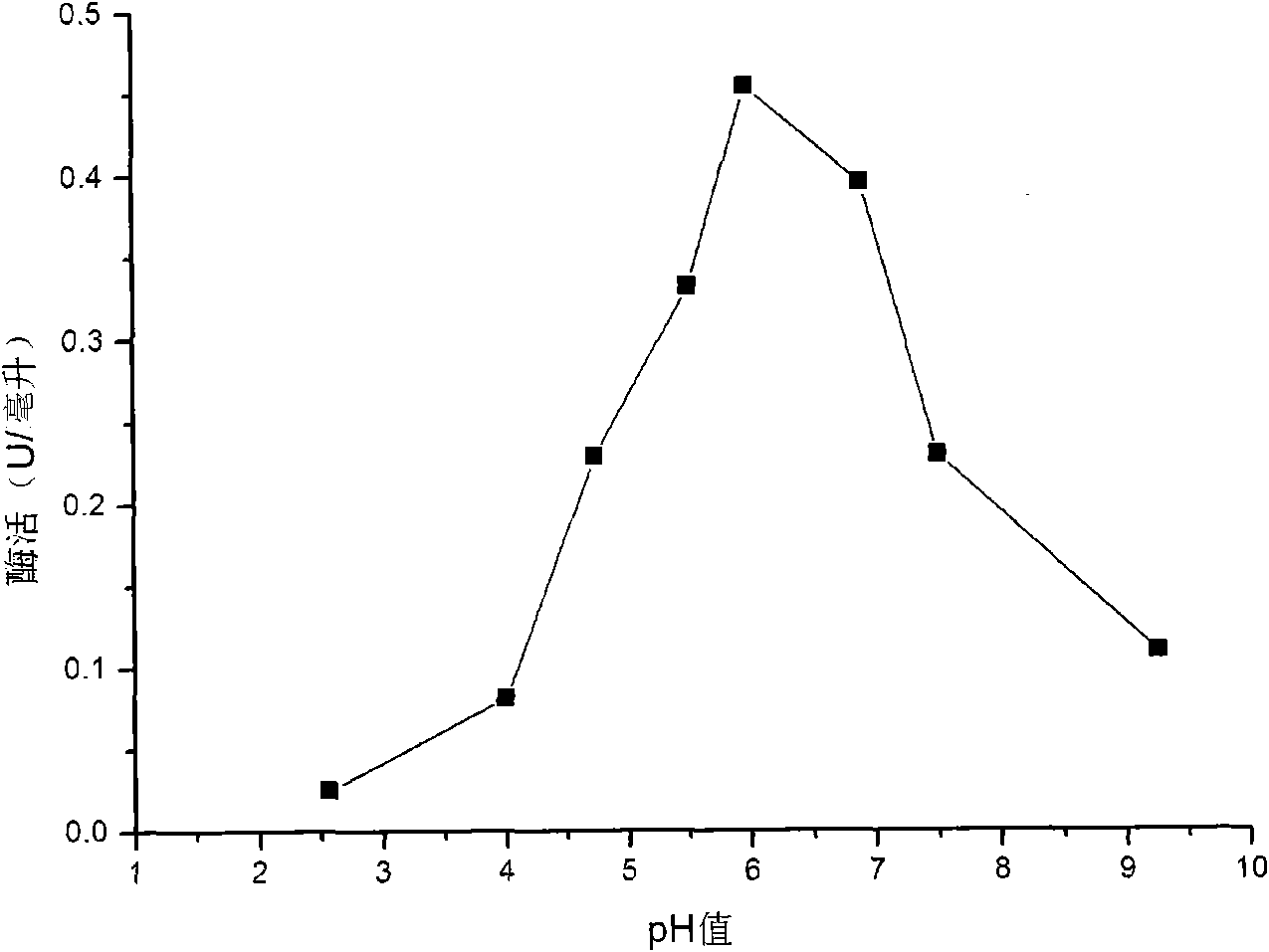
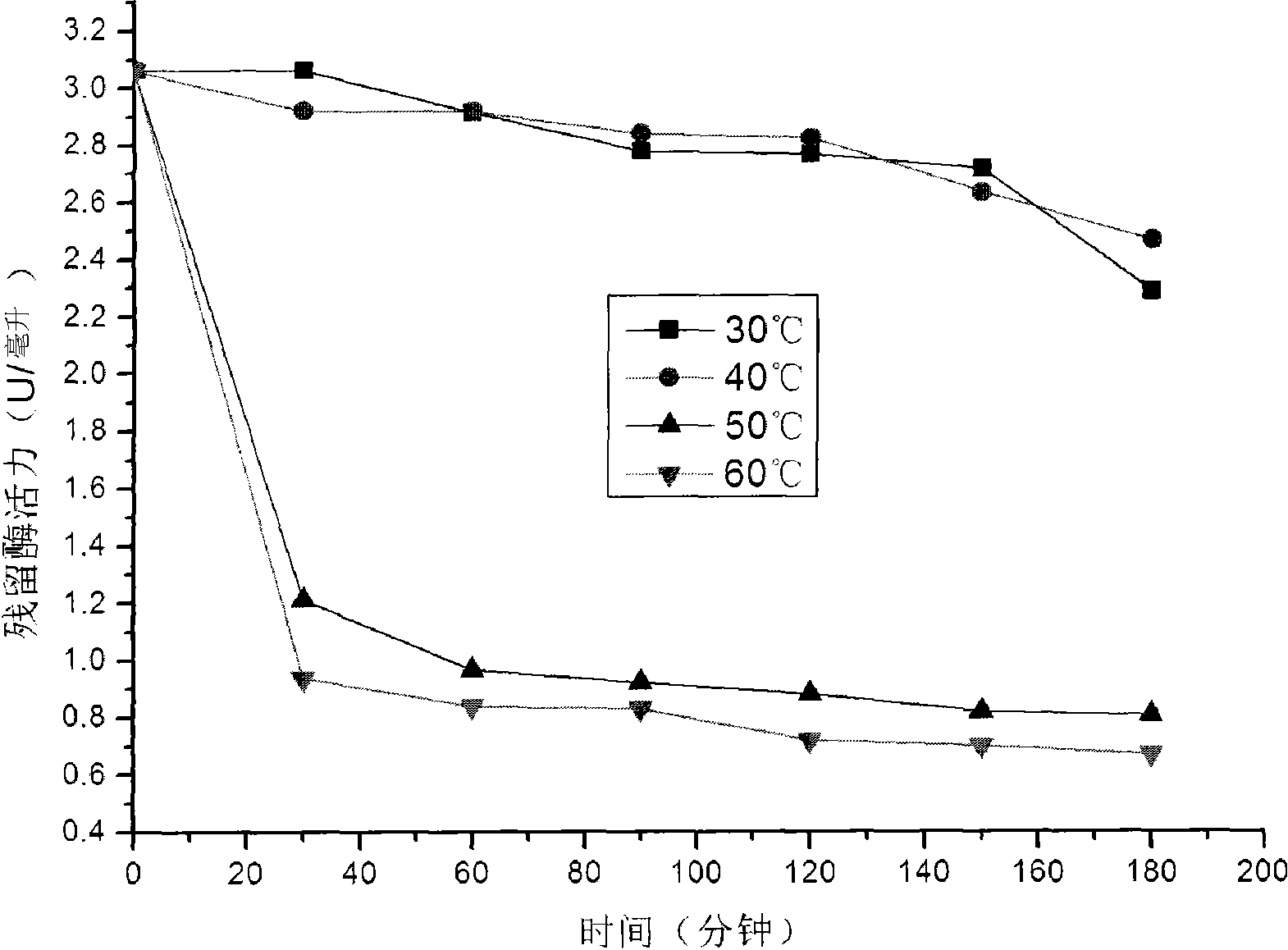
[0043] The obtained crude enzyme liquid takes laminarin as a substrate, and uses 3,5-dinitrosalicylic acid (DNS) method to measure the activity of β-1,3-glucanase, and the β-1,3-glucanase activity of the crude enzyme liquid is - Glucanase activity is 3-5U / ml. The optimum reaction temperature of β-1,3-glucanase is 50°C, and the optimum reaction pH is pH 6. The optimal reaction temperature of β-1,3-glucanase is as follows figure 1 shown. The optimal reaction pH of β-1,3-glucanase is as follows figure 2 s...
Embodiment 3
[0046] Example 3: Preparation of crude enzyme preparation of β-1,3-glucanase
[0047] With the fermented and cultivated crude enzyme liquid of chitosanase producing bacteria H12 obtained in Example 2, centrifuge 30min at 4000-5000rpm, get the supernatant, slowly add solid ammonium sulfate until the concentration of ammonium sulfate is 70-75% (w / v) Saturation, centrifuge at 8000-10000rpm for 30min, put the obtained precipitate into a dialysis bag with a semi-permeable membrane, tie the bag tightly and place it in a 1000mL beaker of 0.01mol / L acetate buffer solution with pH 6.0, dialyze overnight in the refrigerator, and replace About 4 times with acetate buffer. Remove the enzyme solution in the dialysis bag, freeze overnight at -20°C, and freeze-dry in a freeze dryer for 36 hours to obtain a crude enzyme preparation of β-1,3-glucanase in the form of dry powder.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com