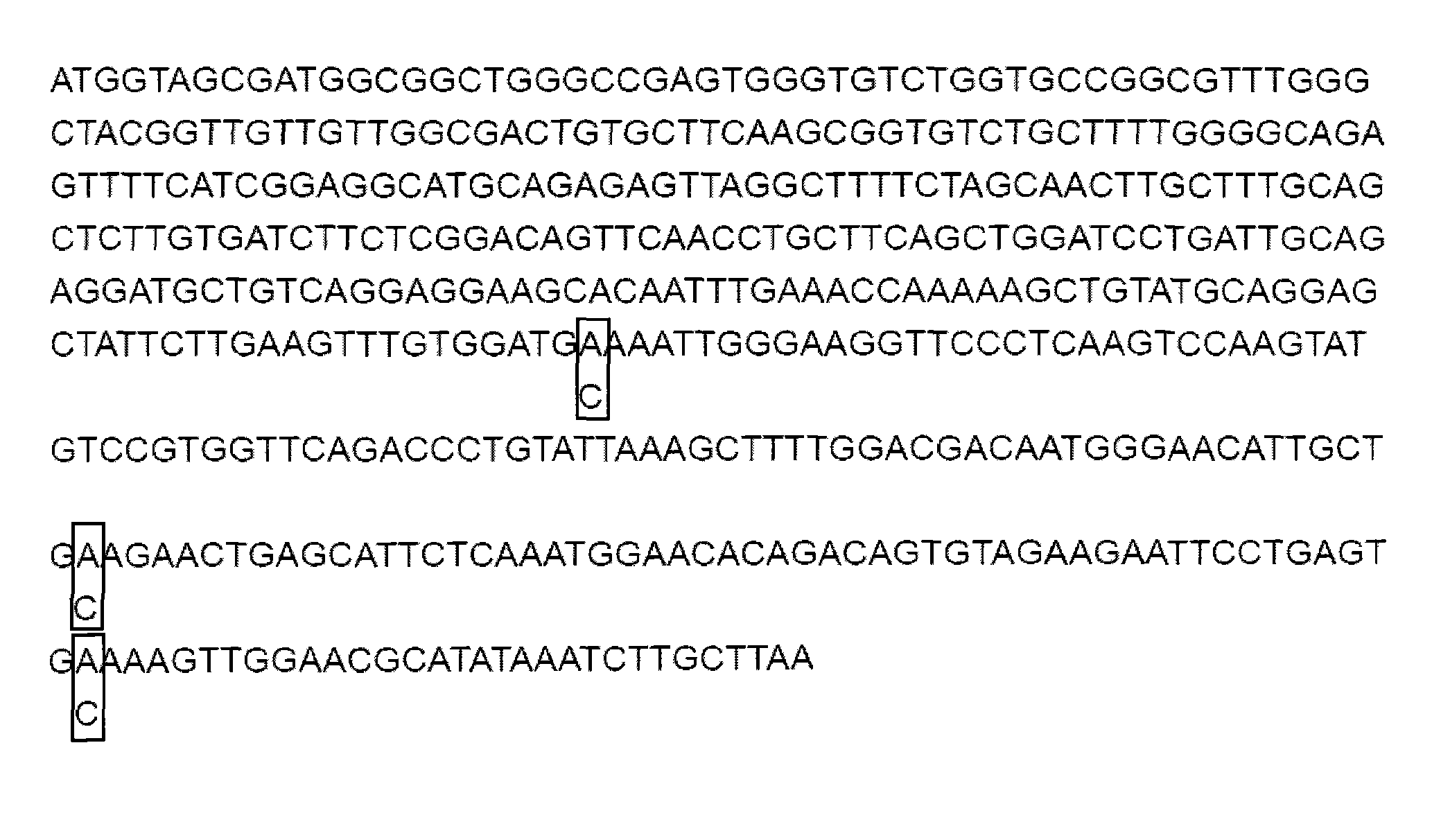Gene site-directed multi-site mutation method
A multi-site mutation and site-specific technology, applied in the field of genetic engineering, can solve the problems of high cost, cumbersome operation and complexity, and achieve the effect of less time and low cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
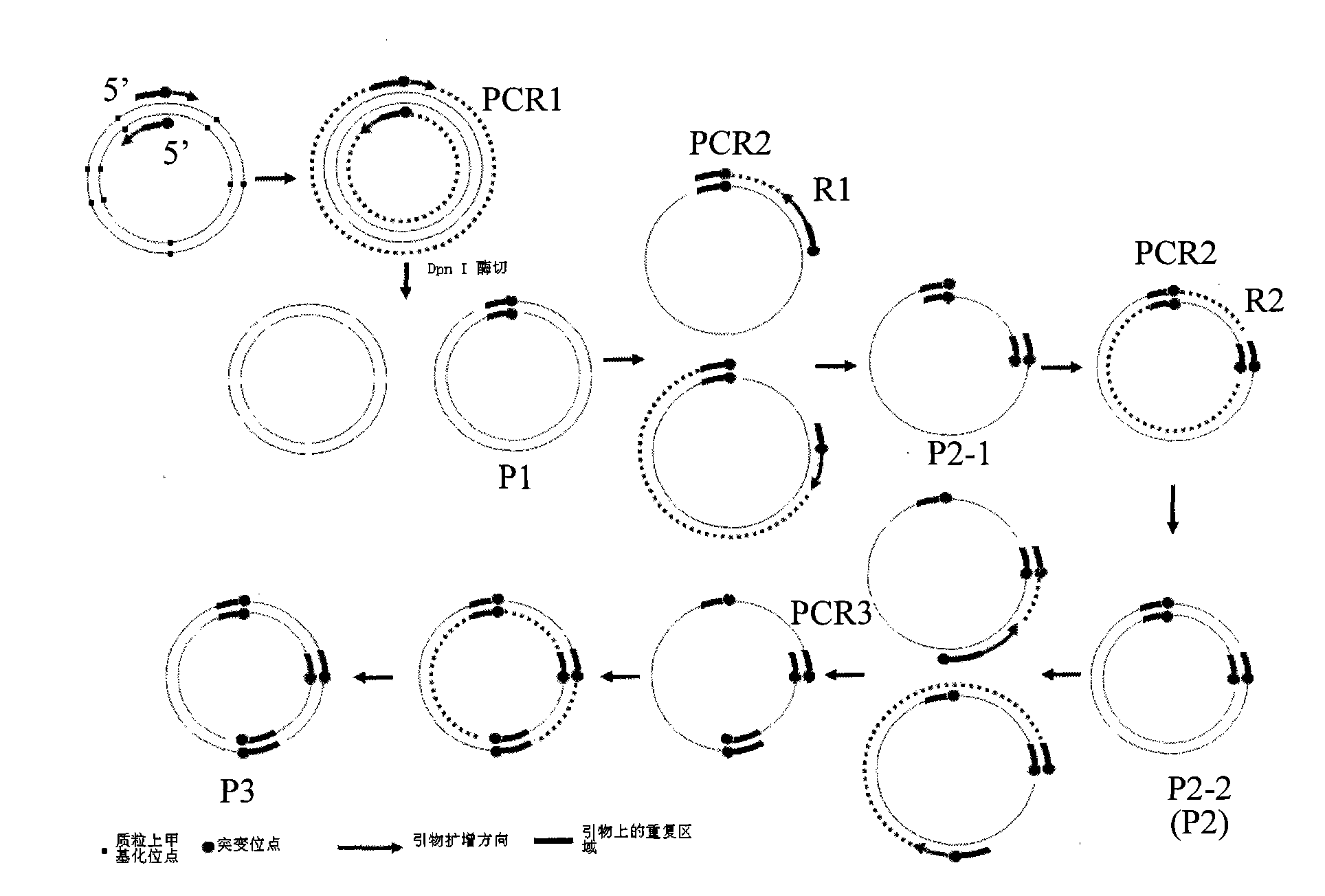
[0048] Embodiment one: 15kDa selenoprotein coding region three site mutations (see attached image 3 ).
[0049] Human 15kDa selenoprotein (Sep 15) wild-type gene has been cloned into pMD18-T (TAKARA) vector, numbered p15-MD-T. The plasmid was extracted from DH5α with the Omega plasmid extraction kit for mutation experiments. The three TGAs on this gene were site-directed mutations to TGCs.
[0050] Sep15 Mutation Primers:
[0051] Sep15mutaF1
5'TTGAAGTTTTGTGGATG C AAATTGGGAAGGTTC 3’
Sep15mutaR1
3'TACGTCCTCGATAAGAACTTCAAACACCTAC G 5'
Sep15mutaF2
5'ATGGGAACATTGCTG C AGAACTGAGCATTCTC 3'
Sep15mutaR2
3'TCGAAAACCTGCTGTTACCCTTGTAACGAC G 5'
Sep15mutaF3
5'TAGAAGAATTCCTGAGTG C AAAGTTGGAACG 3'
Sep15mutaR3
3'TTGTGTCTGTCACATTCTTCTTAAGGACTCAC G 5'
[0052] The first round of PCR:
[0053] PCR was performed with a high-fidelity enzyme (KOD-PLUS, TOYOBO, Japan). The reaction system is as follows: ...
example 2
[0112] Example two: 5 TGAs encoding selenocysteine on the selenoprotein P cDNA are mutated to TGC (see attached Figure 4 )
[0113] The primer list is as follows:
[0114] SelPmutaF1
5'ATCTTTATGTAGCTG C CAGGGACTTCGGGCAG 3'
SelPmutaR1
3'CATTTCTTTTGGAGGGTAGAAATACATCGAC G 5'
SelPmutaF2
5' TG C CGTTTGCCTCCAGCTGCCTG C CAAATAAGTCAGCAGCTTAT 3’
SelPmutaR2
3'GTATTGACTTAGAACAGTCAC G GCAAACGGAGGTCGTCGGAC G 5'
SelPmutaF3
5'AGTGCCAGTTG C CGCTG C AAGAATCAGGC 3’
SelPmutaR3
3'GGTGTCTTCGGTCACGGTCAAC G GCGAC G 5'
[0115] Refer to Example 1 for the PCR and cloning methods.
[0116] Result test:
[0117] The eight clones were sequenced, and the results were as follows: 3 clones caused mutations by all three pairs of primers, and the mutation efficiency was 37.5%; 5 clones caused mutations by both pairs of primers, and the mutation efficiency was 62.5%. Two pairs of primers were induced; the nu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com