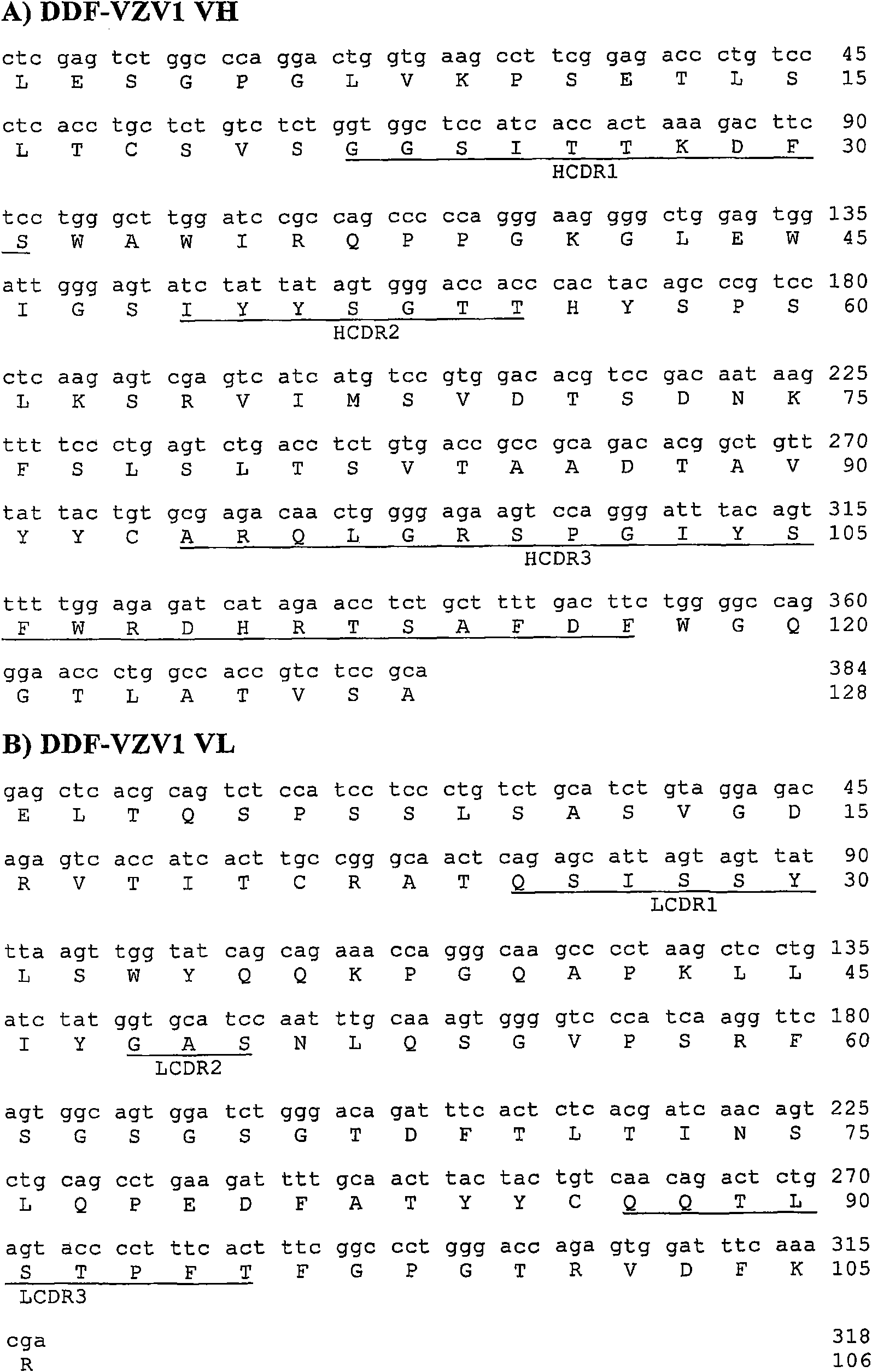Antibodies specific for varicella zoster virus
An antibody and antibody fragment technology, applied in the direction of antibodies, viral peptides, antiviral agents, etc., can solve the problems of non-elimination, reduced treatment, and persistent disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
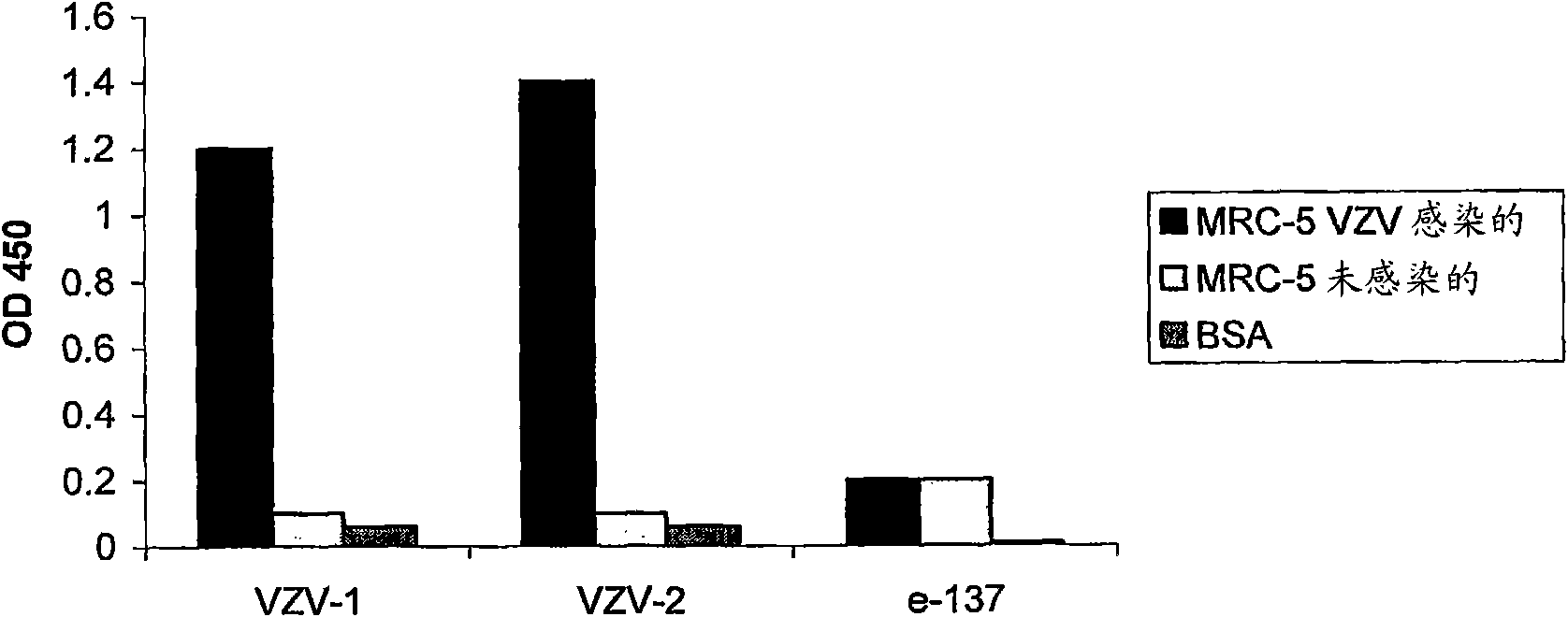
[0091] Example 1: Expression and Selection of Human Fabs Binding VZV Protein Extracts in ELISA
[0092] Materials and methods
[0093] library construction
[0094] According to the literature (Burioni et al., 1998; "Phage Display: A laboratory Manual", Burton DR et al., CSHL Press, 2001), cDNAs encoding the heavy and light chains of human IgG1 were obtained from lymphocytes obtained from From a VZV-seropositive individual. According to the technique described in PCT patent application WO07 / 007154, phage libraries are constructed using cloning cassettes (or cassettes) compatible with pDD vectors, and Fabs are expressed on the surface of recombinant phage in the library.
[0095] Human Fab selection was performed by pDD-based panning (or panning) of the Fab library and sequencing of positive clones as described in the literature (Burioni R et al. 1998).
[0096] The CDRs of specific Fabs were predicted by comparison and sequence alignment provided by IMGT / V-QUEST (Giudicel...
Embodiment 2
[0112] Example 2: Performance of DDF-VZV1 and DDF-VZV2 tested on VZV-infected cell cultures
[0113] Materials and methods
[0114] Neutralization assay of VZV-specific human Fab
[0115] Use 10 4 -10 5 MRC-5 cells were subjected to plaque reduction assays in Costar 24-well plates into which they were plated under conditions in which they generally formed confluent monolayers after 72 hours of incubation at 37°C. The VZV strains used were clinical isolates.
[0116] The Fab was partially purified as indicated in Example 1 and was prepared at various concentrations with the same volume of stock solution of VZV-free cells (ELLEN strain; infection 0.01 multiplicity (or multiplicity)) suspended in maintenance medium (maintenance medium) (0.01, 0.1, 1, 10 and 50 μg / ml) were mixed. Controls consisted of the same volume of maintenance medium and virus in the absence of Fab (blank) or in the presence of an irrelevant Fab specific for human hepatitis C virus (e137; Bugli F et al...
Embodiment 3
[0126] Example 3: Production and confirmation of DDF-VZV1 and DDF-VZV2 using pDD-compatible expression vectors
[0127] Materials and methods
[0128] Design and construction of pDLac-FLAGhis vector
[0129] The pDD vector containing the DDb cassette in which the Zeocine gene is used as a marker gene (WO 07 / 007154) has been modified by replacing the SpeI-NheI fragment including the cp3* sequence with a NheI-SpeI synthetic linker containing two tags and a stop codon. grooming. Such a linker was created by annealing two oligonucleotides (SEQ ID NO: 15 and 16). The resulting double-stranded DNA molecules were digested with SpeI and NheI and prepared for the step of cloning into the corresponding linearized pDD vectors. The final vector was characterized by restriction and sequence analysis to confirm the in-frame insertion of the two tag sequences. The LacI gene was PCT amplified from a commercially available plasmid called pET28 (Invitrogen) and then inserted into the StuI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com