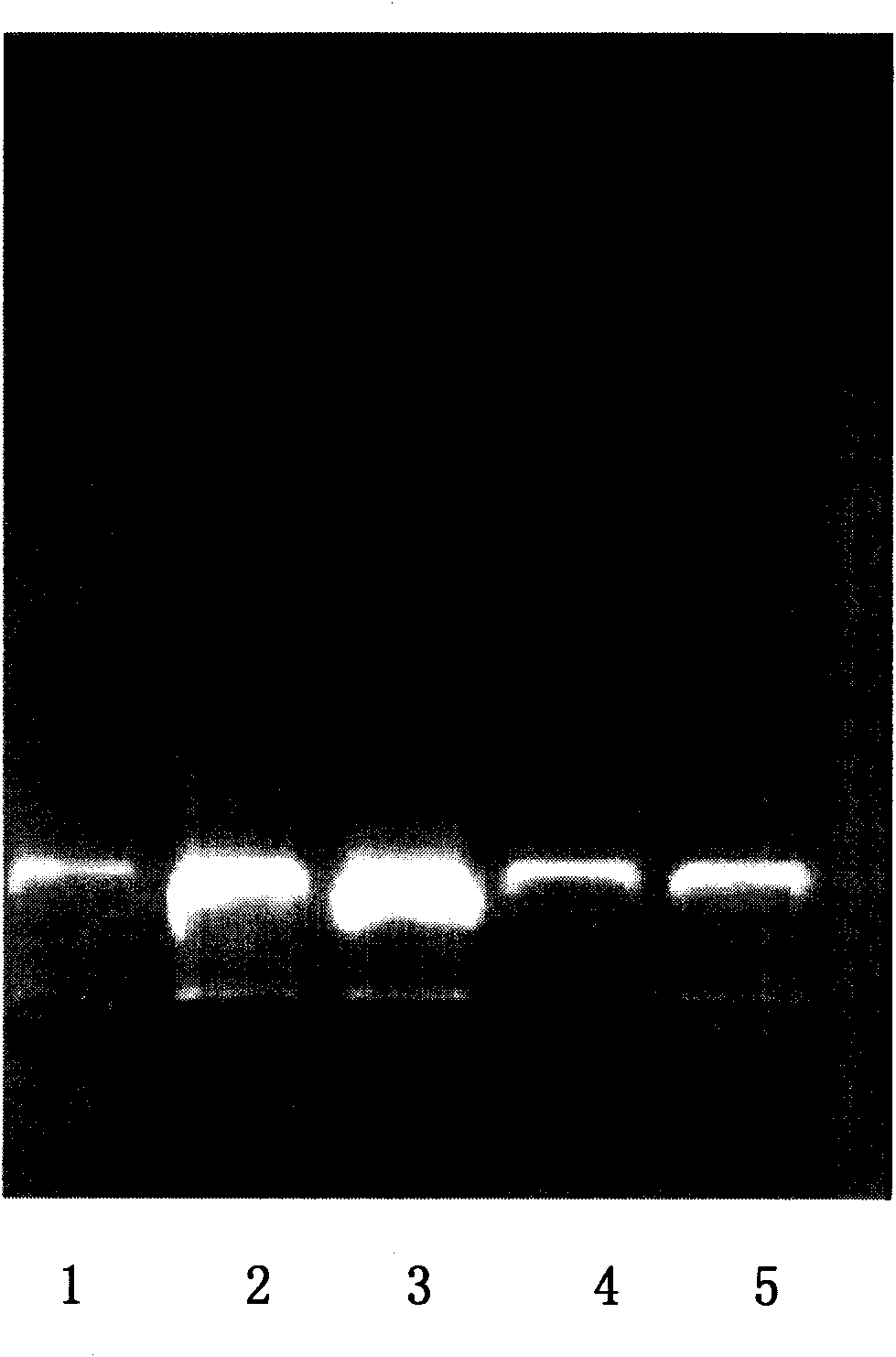Method for extracting total DNA of microorganism in liquor Daqu
A technology of microorganisms and Daqu, applied in DNA preparation, recombinant DNA technology, sugar derivatives, etc., can solve the problems of certain difficulties in extraction methods, large types of microorganisms, and difficulty, and achieve the effect of simple separation and identification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Take 5g Daqu + 25mL phosphate buffer (pH8.0) + 0.1%PVPP, + 1mL soil temperature (80 or 60) shaker for 30min, ultrasonic for 6-7min, let stand for 2-5min, and precipitate at low speed (1000rpm) centrifuge for 5min , take out the supernatant, mix, centrifuge at high speed (9000rpm) for 8min, remove the supernatant, then add phosphate buffer (PVPP), 25mL, break up and centrifuge at high speed (9000rpm) for 8min, remove the supernatant.
[0033] Add 15-25mL extract solution (the components of the extract solution are: 100mM Tris-HCl (tris(hydroxymethyl)aminomethane hydrochloride) pH8.0, 100mM sodium EDTA (disodium ethylenediaminetetraacetate) pH8.0, 100mM Sodium Phosphate pH 8.0, 1.5M NaCl and 1% CTAB (Cetyltrimethylammonium Bromide)), shake well. Add 50ul of 10mg / mL protease K, 37°C, 200rpm, shake horizontally for 30min, shake up and down once in the middle; add 1ml of 50mg / mL lysozyme, add 10ml of 20% SDS after shaking (addition is 4% of the total volume) ), incubated in...
Embodiment 2
[0035] Take 3g Daqu + 25mL phosphate buffer (pH8.0) + 0.05% PVPP, + 1mL Tuwen-60 shaker for 30min, ultrasonic for 6-7min, let stand for 2-5min, centrifuge at low speed (1000rpm) for 5min, remove the supernatant , mix, centrifuge at high speed (9000rpm) for 8min, remove the supernatant, then add phosphate buffer (PVPP), 25mL, break up and centrifuge at high speed (9000rpm) for 8min, remove the supernatant.
[0036] Add 15ml of extract solution (the components of the extract solution are: 100mM Tris-HCl (tris(hydroxymethyl)aminomethane hydrochloride) pH8.0, 100mM sodium EDTA (disodium ethylenediaminetetraacetic acid) pH8.0, 100mM phosphoric acid Sodium pH8.0, 1.5M NaCl and 1% CTAB (cetyltrimethylammonium bromide)) plus 10mg / mL proteinase K 100ul, 37°C, 200rpm, shake horizontally for 30min, shake up and down once in the middle; 1ml of 50mg / mL lysozyme, after shaking, add 3ml of 10% SDS, incubate in a water bath at 50°C for 2 hours, slowly invert the centrifuge tube every 15-20min...
Embodiment 3
[0038] Take 8g Daqu + 25mL phosphate buffer solution (pH8.0) + 0.2% PVPP, + 1mL Tuwen-80 shaker for 30min, ultrasonic for 6-7min, let stand for 2-5min, precipitate at low speed (1000rpm) centrifuge for 5min, take out the supernatant , mix, centrifuge at high speed (9000rpm) for 8min, remove the supernatant, then add phosphate buffer (PVPP), 25mL, break up and centrifuge at high speed (9000rpm) for 8min, remove the supernatant.
[0039]Weigh 5g Daqu and place it in a 50ml centrifuge tube, add 15ml of extract (the extract composition is: 100mM Tris-HCl (tris(hydroxymethyl)aminomethane hydrochloride) pH8.0, 100mM sodium EDTA (ethylene di Disodium amine tetraacetate) pH8.0, 100mM sodium phosphate pH8.0, 1.5M NaCl and 1% CTAB (hexadecyltrimethylammonium bromide)) plus 10mg / mL proteinase K 100ul, 37°C, 200rpm , shake horizontally for 30 minutes, shake up and down once in the middle; add 1ml of 50mg / mL lysozyme, add 3ml of 10% SDS after shaking, incubate in a water bath at 50°C for 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com