Method for rapidly detecting cow and sheep derived components
A sheep source, cattle and sheep technology, applied in the field of molecular biology, to achieve the effect of easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] (1) Reagent: Bst DNA polymerase large fragment produced by BioLabs (NEW ENGLAND) and 10 times Buffer solution; specific primer mixture; 4M betaine solution; 0.2M MgSO 4 solution;
[0050] (2) Amplification reaction system: the total volume of the amplification reaction is 25 μL, and its various components are: 10×Buffer 2.5 μL, 4M betaine 6.25 μL, 0.2M MgSO 4 0.25 μL, 1 μL of mixed primers, 3.5 μL of 10 μM dNTPs, 1 μL of 8000 U / L Bst DNA polymerase large fragment, 1 μL of template DNA, make up to 25 μL with sterilized deionized water, mix well and centrifuge;
[0051] (3) Amplification reaction procedure: proceed at 63°C for 60 minutes, and keep at 80°C for 2 minutes, store at 4°C;
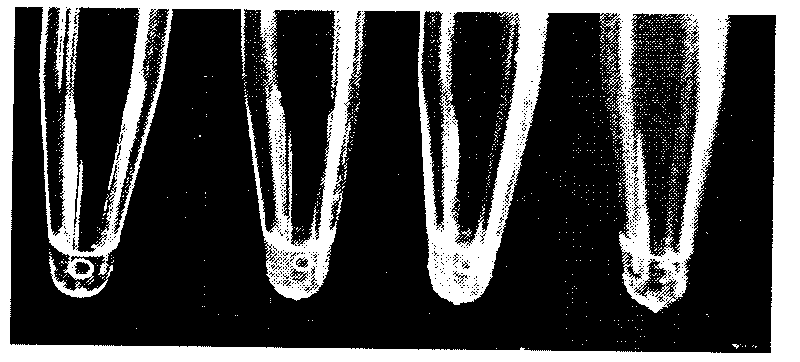
[0052] (4) After the amplification reaction, 15 μL of the system solution was taken, and 1 μL of 1000×SYBRGreen intercalator was directly added to the amplification tube, shaken and mixed, and the results were observed with the naked eye. Tubes without amplification reactions will be ora...
Embodiment 2
[0054] (1) Reagent: Bst DNA polymerase large fragment produced by BioLabs (NEW ENGLAND) and 10 times Buffer solution; specific primer mixture; 4M betaine solution; 0.2M MgSO 4 solution;
[0055] (2) Amplification reaction system: the total volume of the amplification reaction is 25 μL, and its various components are: 10×Buffer 2.5 μL, 4M betaine 6.25 μL, 0.2M MgSO 4 , 0.25 μL of mixed primers, 3.5 μL of 10 μM dNTPs, 2 μL of 8000 U / L Bst DNA polymerase large fragment, 2 μL of template DNA, make up to 25 μL with sterilized deionized water, mix well and centrifuge;
[0056] (3) Amplification reaction procedure: carry out at 65°C for 45min, keep at 80°C for 2min, store at 4°C;
[0057] (4) After the amplification reaction, 15 μL of the system solution was taken, and 2 μL of 1000×SYBRGreen intercalator was directly added to the amplification tube, shaken and mixed, and the results were observed with the naked eye. Tubes without amplification reactions will be orange-yellow, and t...
Embodiment 3
[0059] (1) Reagent: Bst DNA polymerase large fragment produced by BioLabs (NEW ENGLAND) and 10 times Buffer solution; specific primer mixture; 4M betaine solution; 0.2M MgSO 4 solution;
[0060] (2) Amplification reaction system: the total volume of the amplification reaction is 25 μL, and its various components are: 10×Buffer 2.5 μL, 4M betaine 6.25 μL, 0.2M MgSO 4 , 0.25 μL of mixed primers, 3.5 μL of 10 μM dNTPs, 2 μL of 8000 U / L Bst DNA polymerase large fragment, 5 μL of template DNA, make up to 25 μL with sterilized deionized water, mix well and centrifuge;
[0061] (3) Amplification reaction procedure: proceed at 63°C for 45 minutes, and keep at 80°C for 2 minutes, store at 4°C;
[0062] (4) After the amplification reaction was completed, 25 μL of the system solution was taken and analyzed by 2% agarose gel electrophoresis, and the results were observed under ultraviolet light. Tubes without amplification reactions have no obvious bands, and tubes with amplification re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com