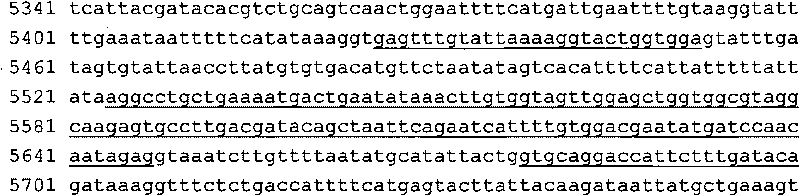Detection method of human proto-oncogene KRAS and kit
A detection method and kit technology, applied in the biological field, can solve problems such as insufficient accuracy, high requirements for experimental conditions, and inability to screen unknown mutations, and achieve the effect of improving accuracy and standardization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1 KRAS gene mutation detection
[0065] Specimen source: This method can be applied to the detection of Kras gene mutations in DNA from blood, tissue, paraffin specimens, pathological sections and other samples. We used DNA extraction from blood, tissue, paraffin and other specimens from Qiagen, Germany. Extraction kit (for specific extraction method, please refer to the kit instruction manual). The samples we selected in this example were tumor tissue samples from 64 cases of colorectal cancer (Fujian Provincial Cancer Hospital, No. 1-64), and DNA was extracted from each tissue.
[0066] PCR amplification:
[0067] Prepare the reaction mixture in a microcentrifuge tube, the reaction system is as follows:
[0068] components
1×
8.5×
wxya 2 o
6.52μl
55.42μl
10×Buffer
1μl
8.5μl
dNTP (10mM)
0.2μl
1.7μl
MgCl 2 (25mM)
0.2μl
1.7μl
Primer Mix (2μM)
1μl
8.5μl
Ta...
Embodiment 2
[0107] Embodiment 2 KRAS gene mutation detection
[0108] The treatment and experimental steps of samples 1-5 are the same as in Example 1. In the PCR amplification step, Primer Mix is a mixture of two pairs of primers, and the concentration of each primer is 2 μM; primer SEQ ID NO: 5 / SEQ ID NO : 6 is a primer for amplifying Kras exon 1; primer SEQ ID NO: 9 / SEQ ID NO: 10 is a primer for amplifying Kras exon 4.
[0109] The sequencing primers in the reaction mixture in the sequencing reaction are KrasE02F and KrasE04F, respectively.
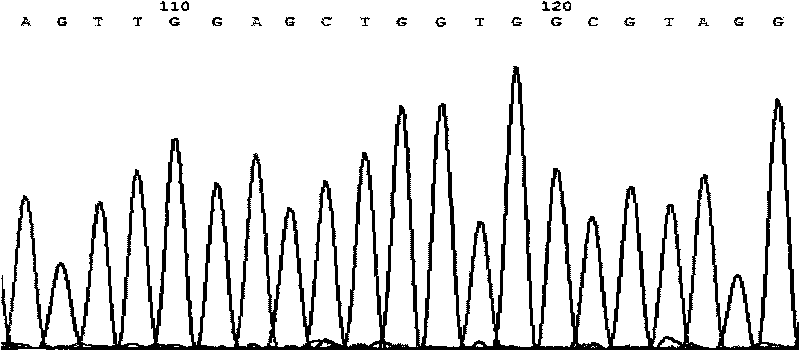
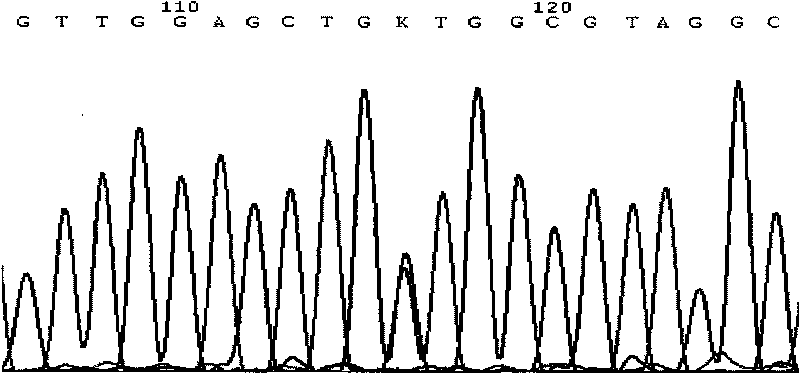
[0110] Sequencing results show that samples 2 and 3 have missense mutations, see image 3 , Figure 4 .
Embodiment 3
[0111] The preparation of embodiment 3KRAS gene mutation detection kit
[0112] Prepare a kit with the following components, each in a tube:
[0113] 1. PCR master mix: Contains 3 pairs of primers (each 2uM), MgCl 2 (25mM), dNTP(10mM), 10×Buffer
[0114] name sequence
Sequence (5'→3')
SEQ ID NO:
KrasE02F: gagtttgtattaaaaggtactggtgga
KrasE03F: cgtcatctttggagcaggaac
KrasE04F: ggaaggaaaatttggtgtagtgg
SEQ ID NO: 5
SEQ ID NO: 7
SEQ ID NO: 9
[0115] name sequence
Sequence (5'→3')
SEQ ID NO:
KrasE02R: gtgcaggacccattctttgataca
KrasE03R: cactgctctaatcccccaaga
KrasE04R: aagaaaccaaagccaaaagca
SEQ ID NO: 6
SEQ ID NO: 8
SEQ ID NO: 10
[0116] 2. Taq DNA polymerase: 1Uhotstar taq enzyme (Germany QIAGEN company)
[0117] 3. PCR product purification enzyme: Contains 0.5U SAP (Promega) and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com