Protein and coding gene and suicide vector thereof
A protein and carrier technology, applied in the field of bioengineering, can solve the problems of multiple screening steps and high false positive rate, and achieve the effects of reducing production costs, improving growth status, and simplifying operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Design and function of embodiment 1 cytotoxic protein gene
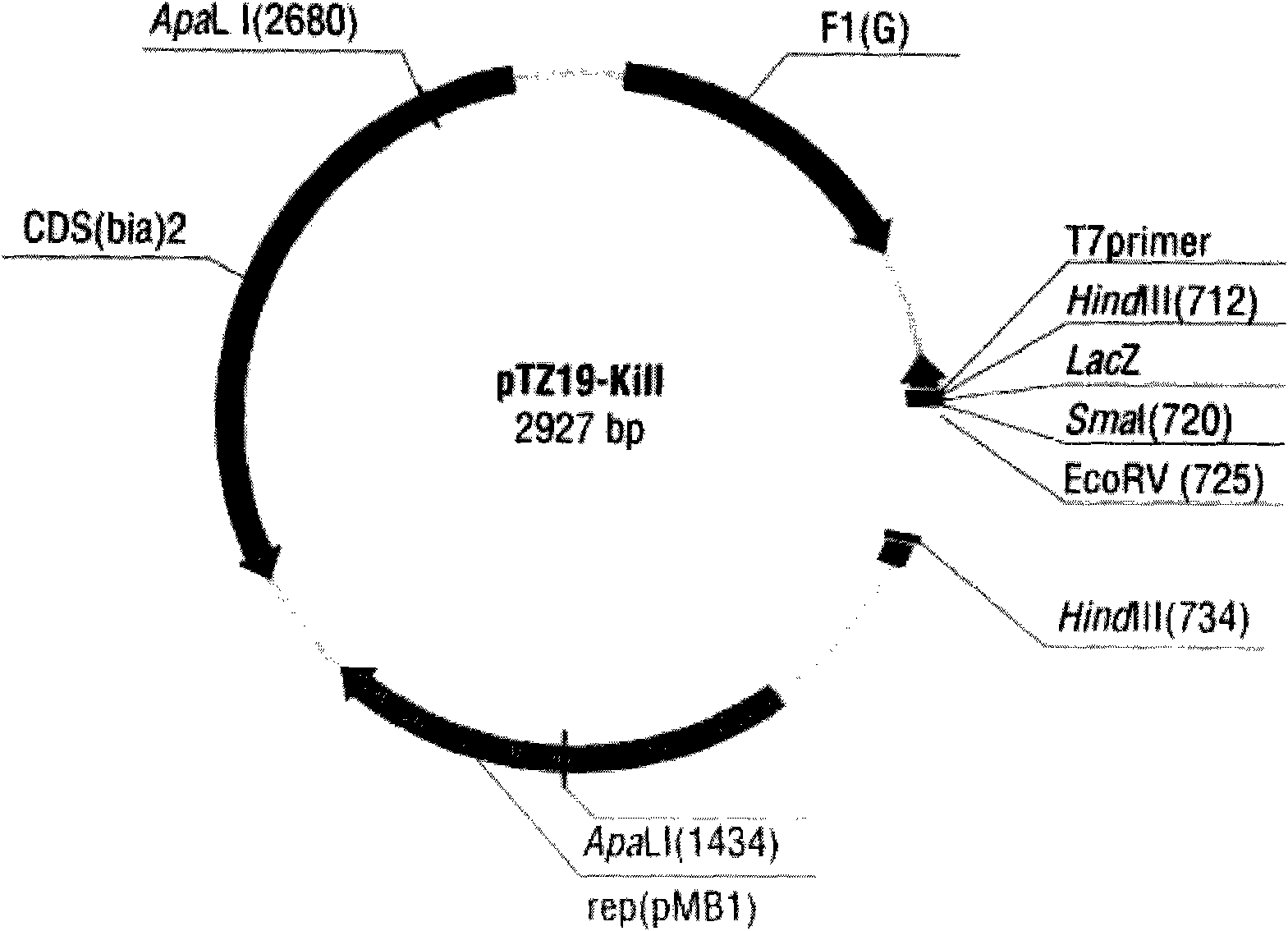
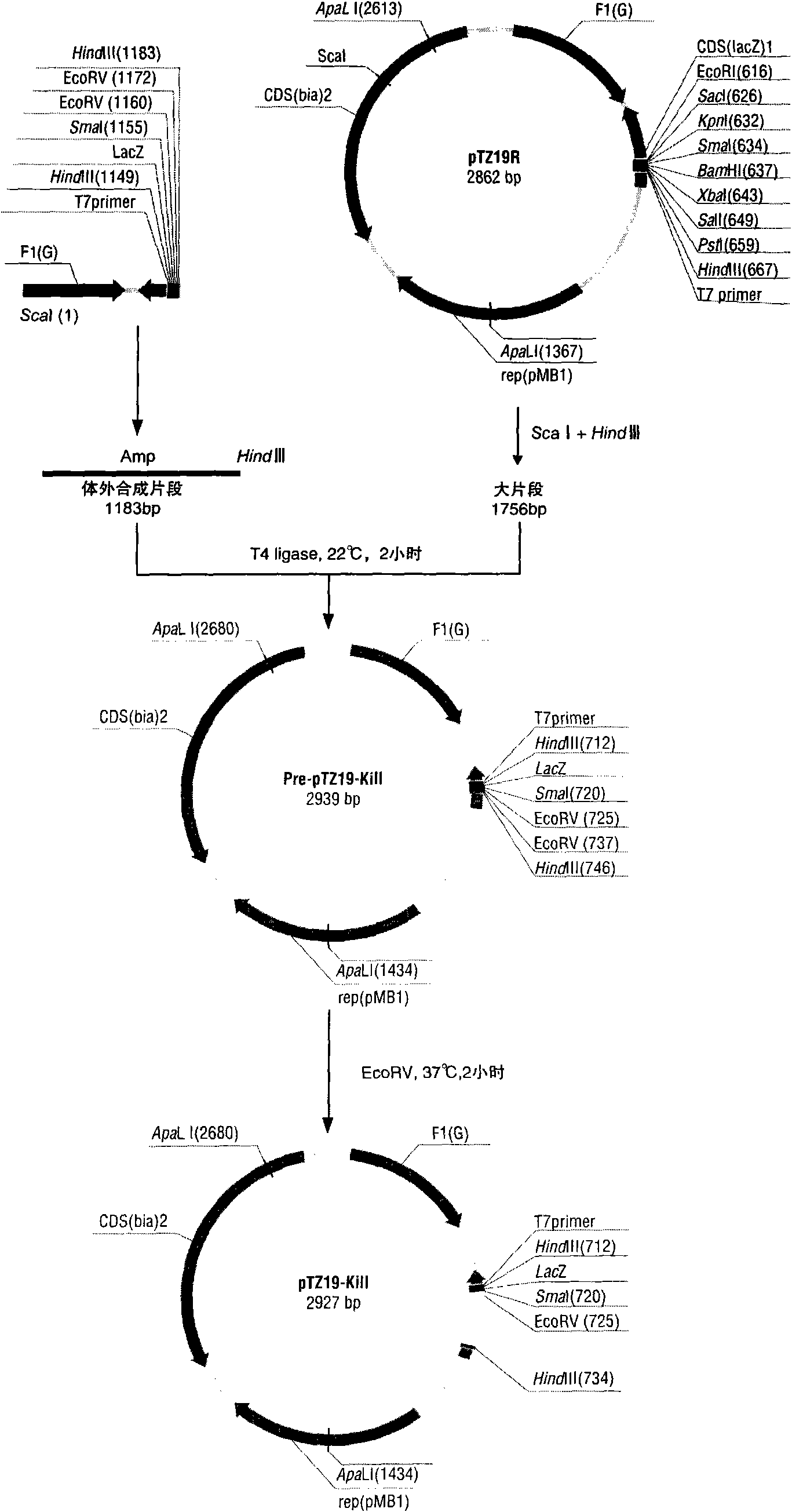
[0024] The inventors unexpectedly discovered that the core functional region of the snake neurotoxic protein whose original coding sequence is ABX58152 is the 37th to 107th positions. Therefore, the inventors further optimized the core functional region and obtained the amino acid sequence of the protein shown in SEQ ID NO: 3 in the sequence listing. Then, according to the codon preference of Escherichia coli, the nucleotide sequence of the amino acid sequence was optimally designed, as shown in the 4th to 267th positions of SEQ ID NO: 2 in the sequence listing. The inventor synthesized the sequence shown in the 4th to 267th positions in SEQ ID NO: 2, cloned it into the plasmid pTZ19R (MBI Fermentas), and transformed the recombinant plasmid into Escherichia coli DH5a competent cells. to grow any transformants.
Embodiment 2
[0025] Example 2 Design and synthesis of β-galactosidase gene-cytotoxic protein gene fusion gene
[0026] The DNA fragment of the nucleotide sequence shown in SEQ ID NO: 2 in the sequence listing was synthesized in vitro by Shanghai Jierui Bioengineering Co., Ltd. by chemical synthesis. Among them, the 4th to 267th positions are the optimized toxic protein coding sequence of the present invention. Positions 268-552 are β-galactosidase genes including multiple cloning sites and blocking sequences. Positions 468-508 are multiple cloning sites, and positions 483-494 are an exogenous blocking fragment. The foreign fragment blocks the reading frame of the fusion gene of β-galactosidase gene-cytotoxic protein gene-multicloning site.
Embodiment 3
[0027] Example 3 Construction of suicide vector
[0028] Carry out following polymerase chain reaction (PCR) respectively: With the DNA fragment synthesized in embodiment 1 as template, with following primer F and R1 as primer pair; With plasmid pTZ19R (MBI Fermentas) as template, with following primer F1 and R is a primer pair; then using the above two amplification products as templates, using primers F and R as a primer pair, the resulting PCR product contains a ScaI (agtact) restriction site at the 5'-end and a HindIII (agtact) at the 3'-end. aagctt) restriction site fragment, spare.
[0029] The primer sequences used in the PCR reaction were:
[0030] Primer F: 5'-agtactcaaccaagtcattctgagaatagtgtat-3',
[0031]Primer R1: 5'-atcatggatcacccatgaagggtgatggttcacgtagtg-3',
[0032] Primer F1: 5'-cactacgtgaaccatcacccttcatgggtgatccatgata-3',
[0033] Primer R: 5'-aagcttcacgtgatatcgcctggg-3'.
[0034] The PCR reaction system is:
[0035] template DNA
1μl
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com