Transcriptional control DNA in vitro transcription
A technology of in vitro transcription and sequence, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The construction of embodiment 1 gene in vitro transcription vector
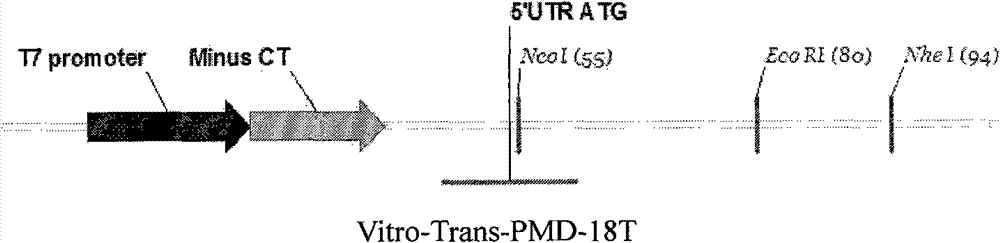
[0037] 1 Determination of the sequence: According to the conditions for the gene to be transcribed into highly translatable mRNA in vitro, we selected the T7 promoter to initiate transcription; in addition, based on the literature and years of experience, we designed a 14bp CT sequence to enhance transcription efficiency; finally, we inserted the gene A sequence containing an ATG initiation site was designed at the enzyme cleavage site to initiate mRNA synthesis. See Figure 2 for the sequence: Figure 2A .In vitro transcription vector map; Figure 2B .In vitro transcription vector sequence 2 Primer design, overlap extension PCR and connection with T vector: According to the designed sequence, we designed 2 pairs of overlapping primers, of which P1 and P2 overlapped by 10bp, each containing 35 bases. P3 and P4 overlap with the products of P1 and P2 by 7 bases, each containing 30 bases. The annealing...
Embodiment 2
[0038] Example 2 The control gene EGFP and the acquisition of 6 key gene cDNAs for maintaining stem cell self-renewal
[0039] 1 Sequence determination and primer design: query EGFP and porcine Oct4, Sox2, c-Myc, Klf4, Nanog, Lin28 mRNA sequences in the NCBI database, and then design PCR primers to amplify their sequences. Oct4 (NM_001113060.1), Sox2 (NM_001123197.1), c-Myc (NM_001005154.1), Klf4 (NM_001031782.1), Nanog (NM_001129971.1) and Lin28 (NM_001123133.1). Primers were designed according to the sequence obtained from the query and the optimal principle of correct expression. Both ends of the primers have endonuclease sites (NcoI, EcoRI and NheI) for connecting to the in vitro transcription vector.
[0040] 2RT-PCR and connection with T carrier: Embryos about 20 days old were washed out from pregnant sows, ground with liquid nitrogen, and then RNA was extracted, then reverse transcribed, and then reverse transcribed cDNA was used as a template for 6 PCR assays. The PC...
Embodiment 37
[0041] The connection of embodiment 37 genes and in vitro transcription carrier
[0042]Enzyme digestion of 17 genes and in vitro transcription vectors: Oct4-T plasmids were digested with EcoRI and NheI, Sox2-T, c-Myc-T, Klf4-T, Nanog-T and Line8-T plasmids were digested with NcoI and NheI . Then electrophoresis, gel cutting, and recovery. The in vitro transcription vector is also digested with corresponding enzymes at the same time. Then, the enzyme-digested fragments of the 7 genes were connected to the in vitro transcription vector, transformed, and shaken for identification.
[0043] Linearization of in vitro transcription vectors for 27 genes: The in vitro transcription plasmids of 7 correctly identified genes were digested with NheI to linearize them to facilitate in vitro transcription.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com