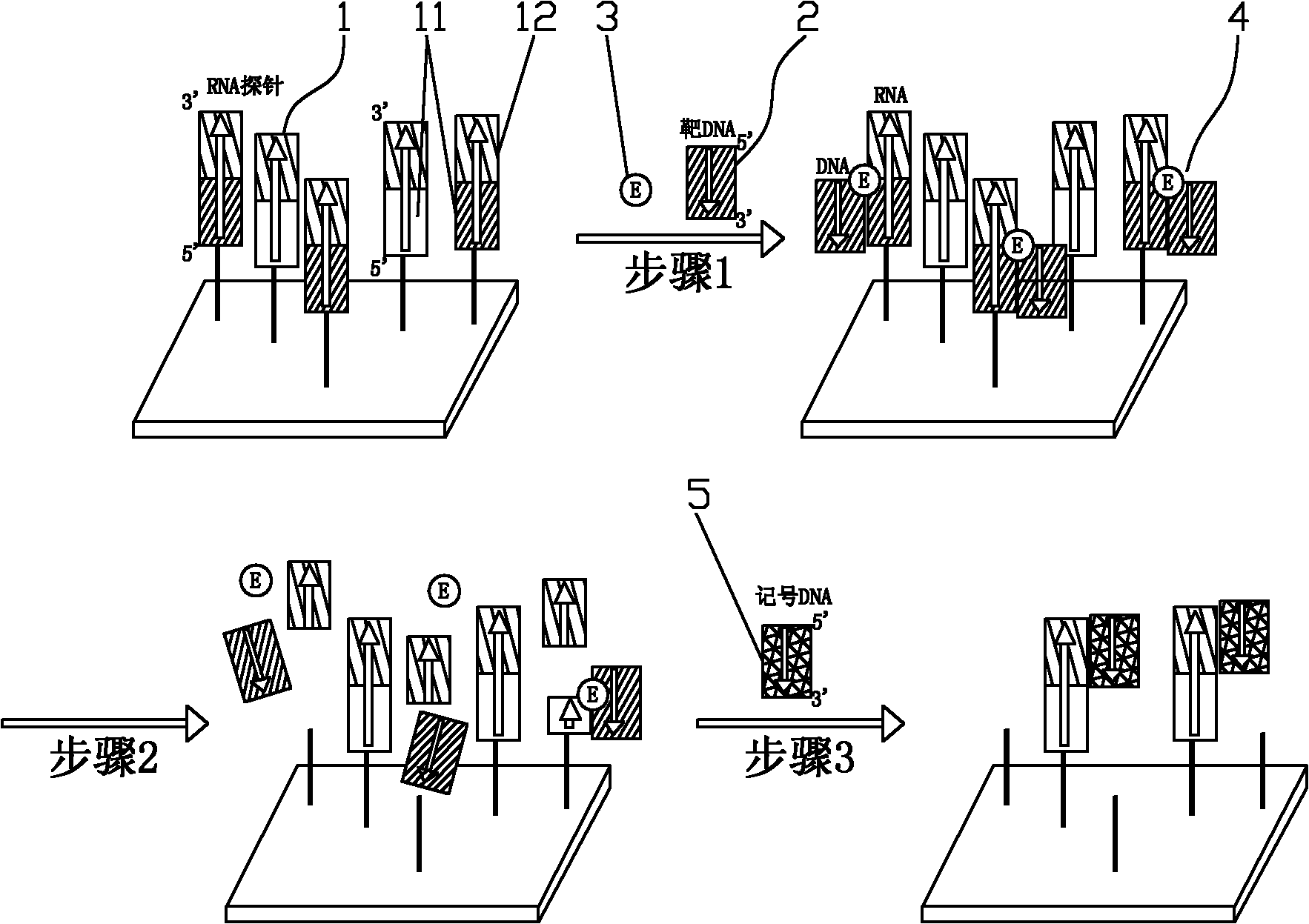
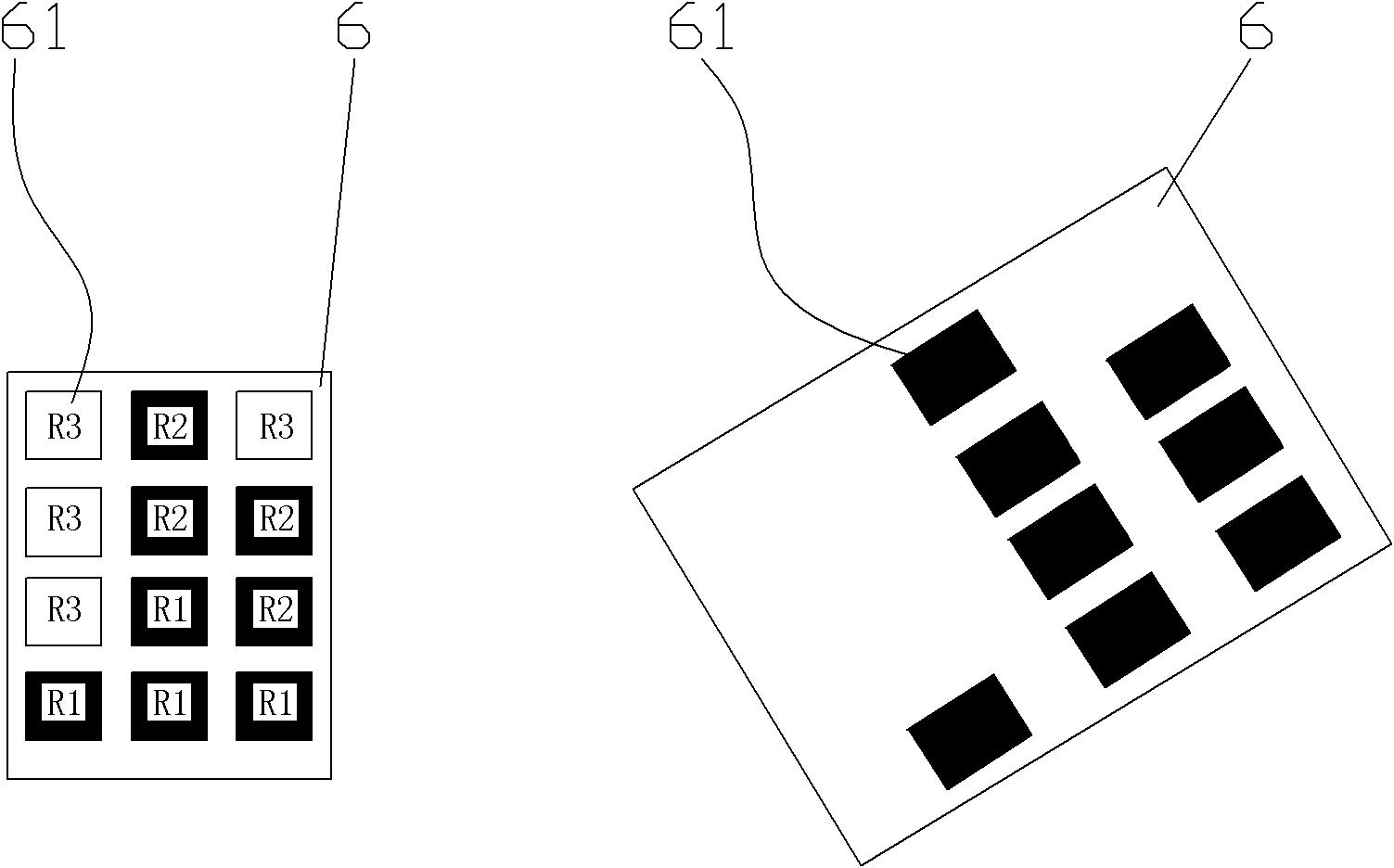
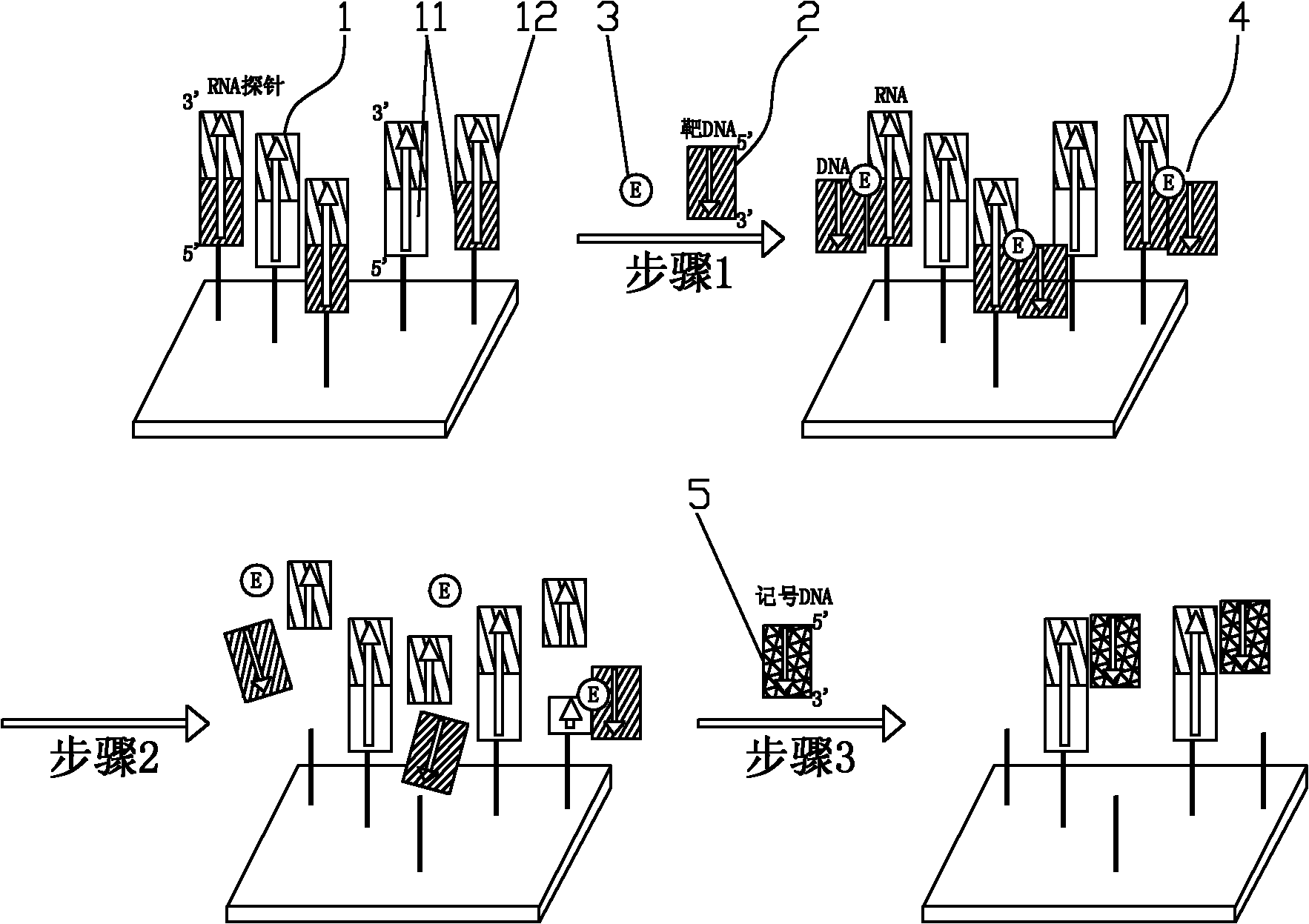
Method for detecting target DNA sequence, gene chip adopting same and application of gene chip
A DNA sequence and gene chip technology, applied in the application field of the method, can solve the problems of amplification pollution, difficult to meet the detection requirements, false negatives, etc., to reduce nucleic acid pollution, improve the accuracy of detection, and avoid false negatives and false positives The effect of the result
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The present embodiment uses the gene chip of the present invention to detect Shigella, wherein:
[0042]The target DNA is from the genome of Shigella, which is double-stranded DNA and consists of SEQ ID NO: 1;
[0043] The RNA probe is artificially synthesized and consists of SEQ ID NO: 2. The sequence shown in the 1-22 positions of SEQ ID NO: 2 is the detection sequence, and the sequence shown in the 23-42 positions is the marker sequence; the detection of the RNA probe The sequence is identical to the sequence shown in positions 478-499 of SEQ ID NO: 1, and is complementary to the complementary strand of the sequence;
[0044] Marked DNA is artificially synthesized and consists of SEQ ID NO: 3, in which the 5' end is labeled with biotin;
[0045] The specific experimental method is as follows:
[0046] 1. Genomic DNA extraction;
[0047] Transfer 1.5ml of the culture into a centrifuge tube and centrifuge at 12,000×g for 3min;
[0048] Discard the supernatant, susp...
Embodiment 2
[0066] In this embodiment, the gene chip of the present invention is used to detect the DNA of the transgenic component Cry1A(b) of the transgenic corn Bt11, wherein:
[0067] The target DNA is from transgenic corn Bt11, which is double-stranded DNA and consists of SEQ ID NO: 5;
[0068] The RNA probe is artificially synthesized and consists of SEQ ID NO: 6. The sequence shown in the 1-21 positions of SEQ ID NO: 6 is the detection sequence, and the sequence shown in the 22-41 positions is the marker sequence; the detection of the RNA probe The sequence is identical to the sequence shown in the 214-234 positions of SEQ ID NO: 1, and is complementary to the complementary strand of the sequence;
[0069] The marked DNA is artificially synthesized and consists of SEQ ID NO: 7, in which the 5' end is labeled with biotin;
[0070] The specific experimental method is as follows:
[0071] 1. Genomic DNA extraction: (extract composition: Tris-HCl pH7.5, 150mM; NaCl 100mM; EDTA pH8.0,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com