RNA in situ hybridization
An in situ hybridization and oligonucleotide technology, applied in the field of RNA in situ hybridization, can solve the problems of increasing background noise and high permeability of tissue samples, and achieve the effect of improving diagnostic accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
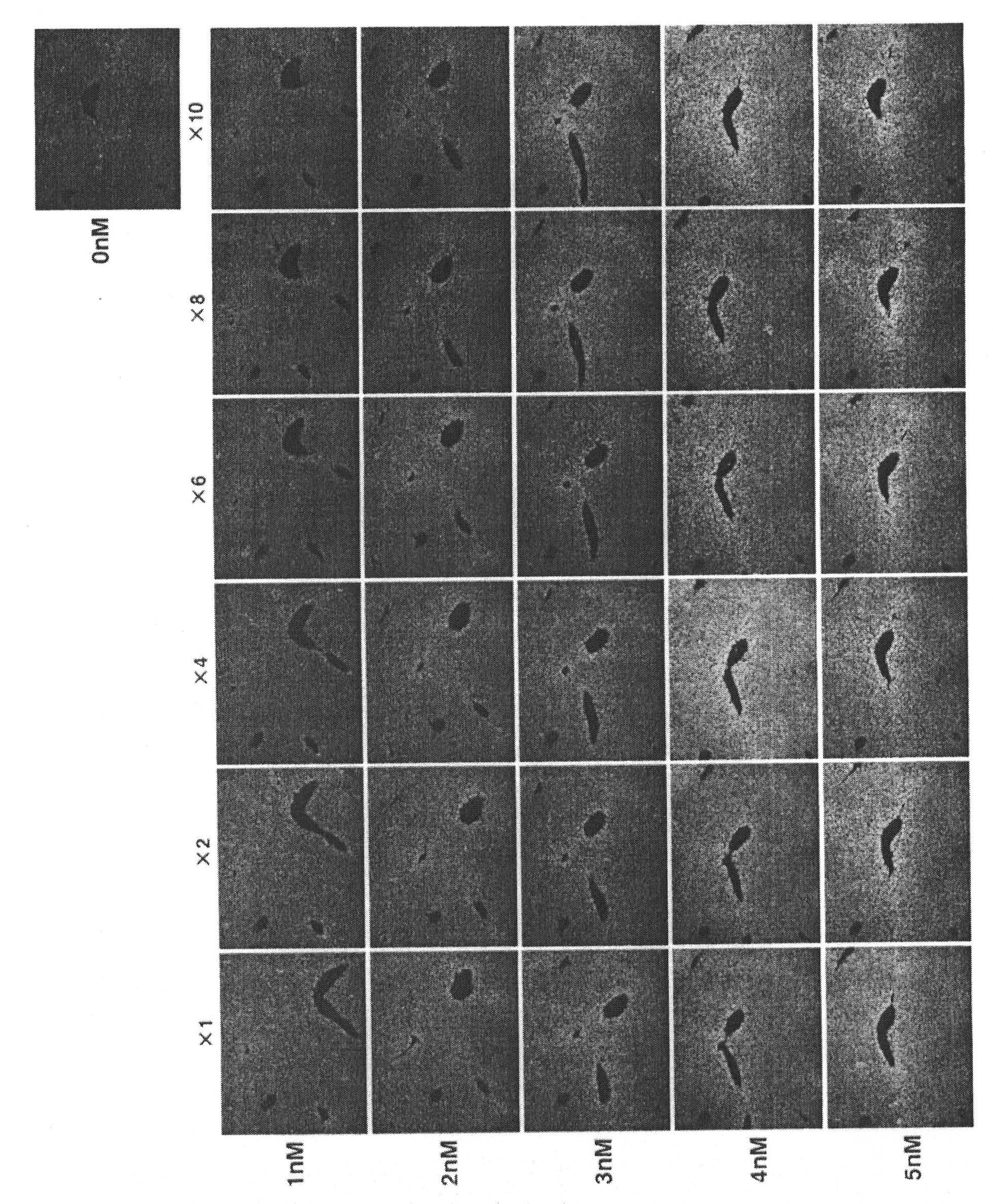
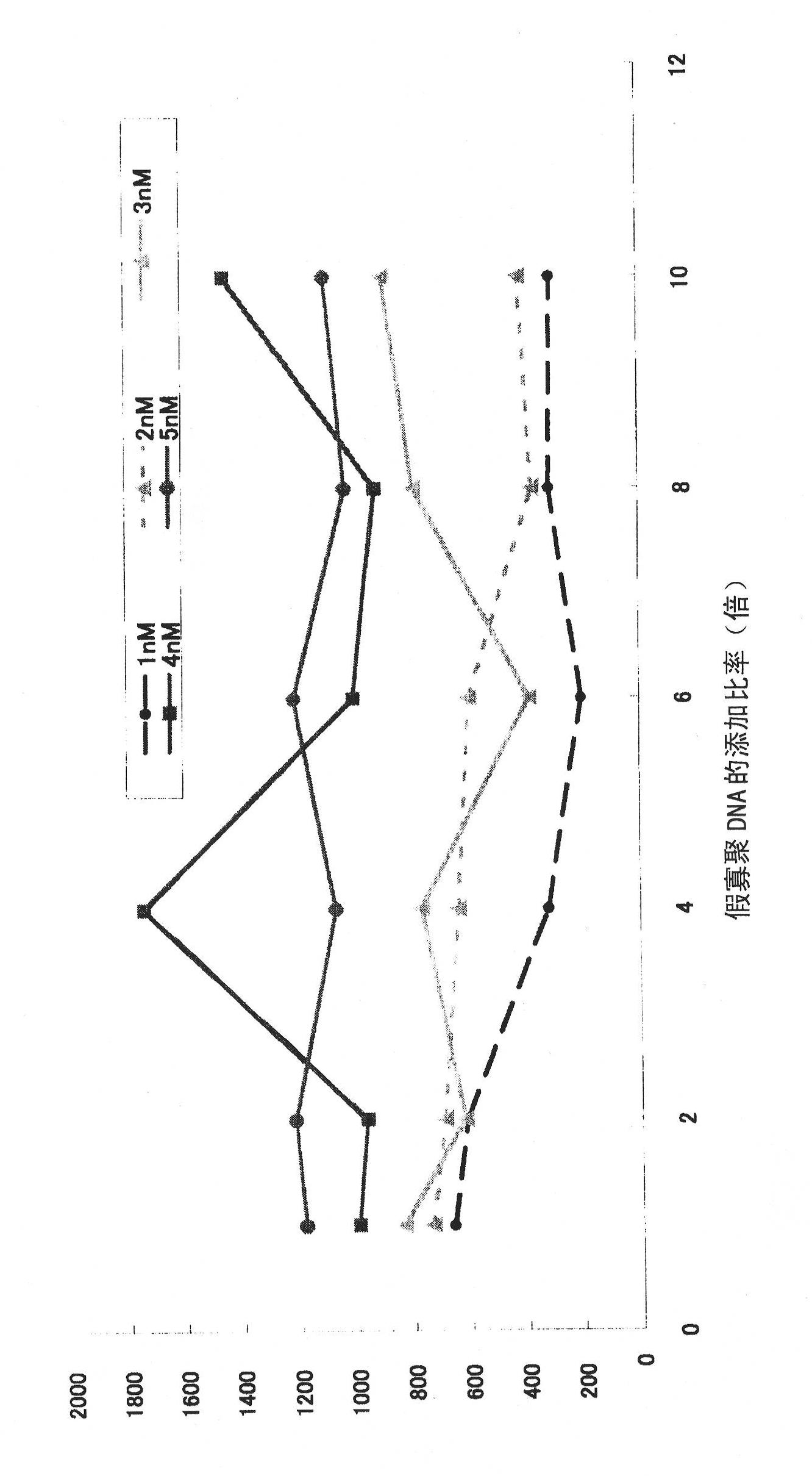
[0153] Embodiment 1 (the experiment of false oligonucleotide addition ratio)
[0154] The optimum addition ratio corresponding to the probe concentration of the dummy oligo DNA was obtained. Mouse liver was used as the target tissue, and the gene to be detected was Cyp1a2. The livers of 8-week-old male mice were usually fixed in formalin and embedded in paraffin to make paraffin blocks, made into serial sections with a thickness of 5 μm, and deparaffinized, and then proteinase K (Invitrogen Company, Proteinase K SOL.RNA, 25530049) treatment, followed by RNA in situ hybridization. As probes for detecting mRNA of the gene Cyp1a2, four types of single-stranded oligo DNA probes (SEQ ID NOS: 1 to 4) whose both ends were labeled with FITC were used. On the mRNA of the Cyp1a2 gene, sequence numbers 1 to 4 are arranged in the order of numbering along the direction from the 5' end to the 3' end, and the distance to the adjacent oligo DNA probe is 594 for sequence numbers 1 and 2 Bas...
Embodiment 2
[0155] Embodiment 2 (false oligomeric DNA, salmon sperm DNA and the comparison without adding)
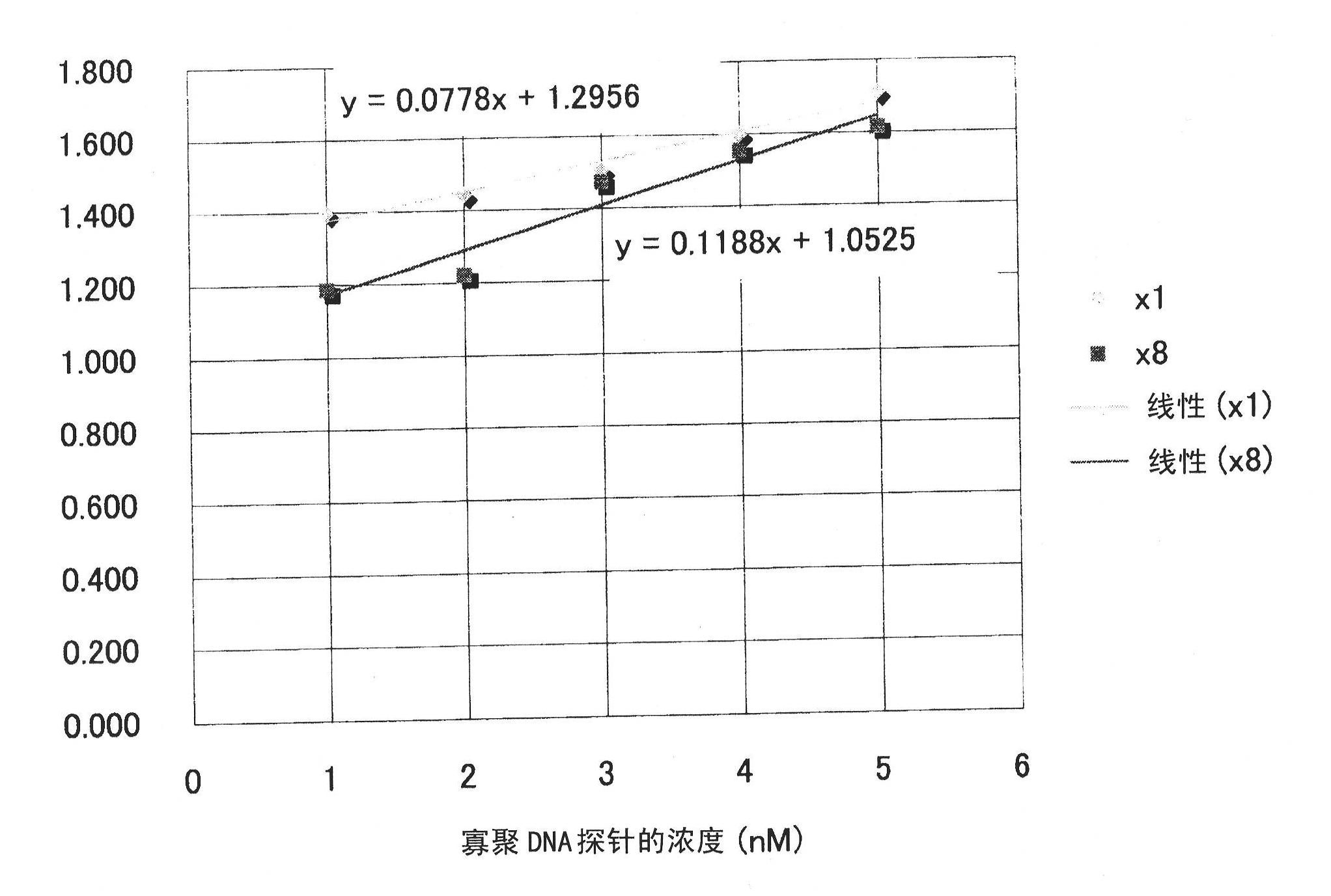
[0156] The effects of pseudo-oligo DNA, salmon sperm DNA, and quantitative RNA in situ hybridization RNA detection without addition were compared. In the experiment, a formalin-fixed and paraffin-embedded mouse liver tissue sample was used in the same manner as in Example 1, and serial sections were prepared and used in the experiment. The detection gene is the same as in Example 1, using Cyp1a2 and FITC-labeled 4 kinds of single-stranded oligomeric DNA probes shown in sequence numbers 1 to 4 at both ends, and the respective concentrations of the 4 kinds of probes are 0 nM (nano mol), 1nM, 2nM, 3nM, 4nM, and 5nM, corresponding to various probe concentrations, and the concentration shown in SEQ ID NO: 5 (denoted as L1C1) and SEQ ID NO: 6 (denoted as arbp) was used at a concentration that was 8 times the addition ratio. 2 types of pseudo-oligomeric DNA (single-stranded). In additio...
Embodiment 3
[0157] Embodiment 3 (comparison of false oligomeric DNA and salmon sperm DNA)
[0158]The effects of quantitative RNA in situ hybridization on false oligo DNA and salmon sperm DNA in RNA detection were compared. In the experiment, serial sections were prepared from formalin-fixed and paraffin-embedded mouse liver tissue samples in the same manner as in Example 1, and used in the experiment. The detection gene is the same as in Example 1, using Cyp1a2 and FITC-labeled 4 kinds of single-stranded oligomeric DNA probes shown in sequence numbers 1 to 4 at both ends, and the respective concentrations of the 4 kinds of probes are 0 nM (nano mol), 1 nM, 2 nM, 3 nM, 4 nM, and 5 nM of various probe concentrations, the pseudo-oligo DNA (single-stranded) shown in SEQ ID NO: 5 was used at a concentration of 8 times the addition ratio. In addition, salmon sperm DNA from Invitrogen (catalogue number 15632-011, Salmon Sperm DNA solution) was added to a final concentration of 100 ug / ml (corre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com