Primer used for detecting c-kit gene mutation and application thereof
A genome and primer sequence technology, applied to the primers used to detect c-kit gene mutation and its application field, can solve the problems of slow disease progression and prolongation of average survival period, and achieve the effect of high reference significance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 A method for identifying mutations in exon 9 of kit
[0031] 1. Extract patient tumor tissue or blood genomic DNA;
[0032] 2. Carry out PCR amplification. The 20 μl PCR reaction system is as follows: 50-100 ng of tumor tissue or blood genomic DNA, 0.125 μl of Taq enzyme, 1 μl of upstream and downstream primers (10 μM), 2 μl of dNTP (2.5 mM), 10×PCR buffer solution (containing Mg2+) 2 μl, and the rest was sterile distilled water; PCR reaction conditions were: denaturation at 95°C for 2 min, followed by denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 1 min, 45 cycles, and finally extension at 72°C for 7 min. Store at 4°C;
[0033] 3. Gel electrophoresis analysis of PCR amplification products:
[0034] a. Rinse the electrophoresis tank and comb with distilled water. Put it on a level table and set up the comb. 1.5% agarose can be prepared for electrophoresis.
[0035] b. Put 50ml of 1×TAE electrophoresis buffer and 100ml...
Embodiment 2
[0054] Example 2 A method for identifying mutations in exon 11 of kit
[0055] 1. Extract patient tumor tissue or blood genomic DNA;
[0056] 2. Carry out PCR amplification. The 20 μl PCR reaction system is as follows: 50-100 ng of tumor tissue or blood genomic DNA, 0.125 μl of Taq enzyme, 1 μl of upstream and downstream primers (10 μM), 2 μl of dNTP (2.5 mM), 10×PCR buffer solution (containing Mg2+) 2 μl, and the rest was sterile distilled water; PCR reaction conditions were: denaturation at 95°C for 2 min, followed by denaturation at 95°C for 30 sec, annealing at 64°C for 30 sec, extension at 72°C for 1 min, 45 cycles, and finally extension at 72°C for 7 min. Store at 4°C;
[0057] 3. Gel electrophoresis analysis of the PCR amplification product, the specific steps are the same as in Example 1.
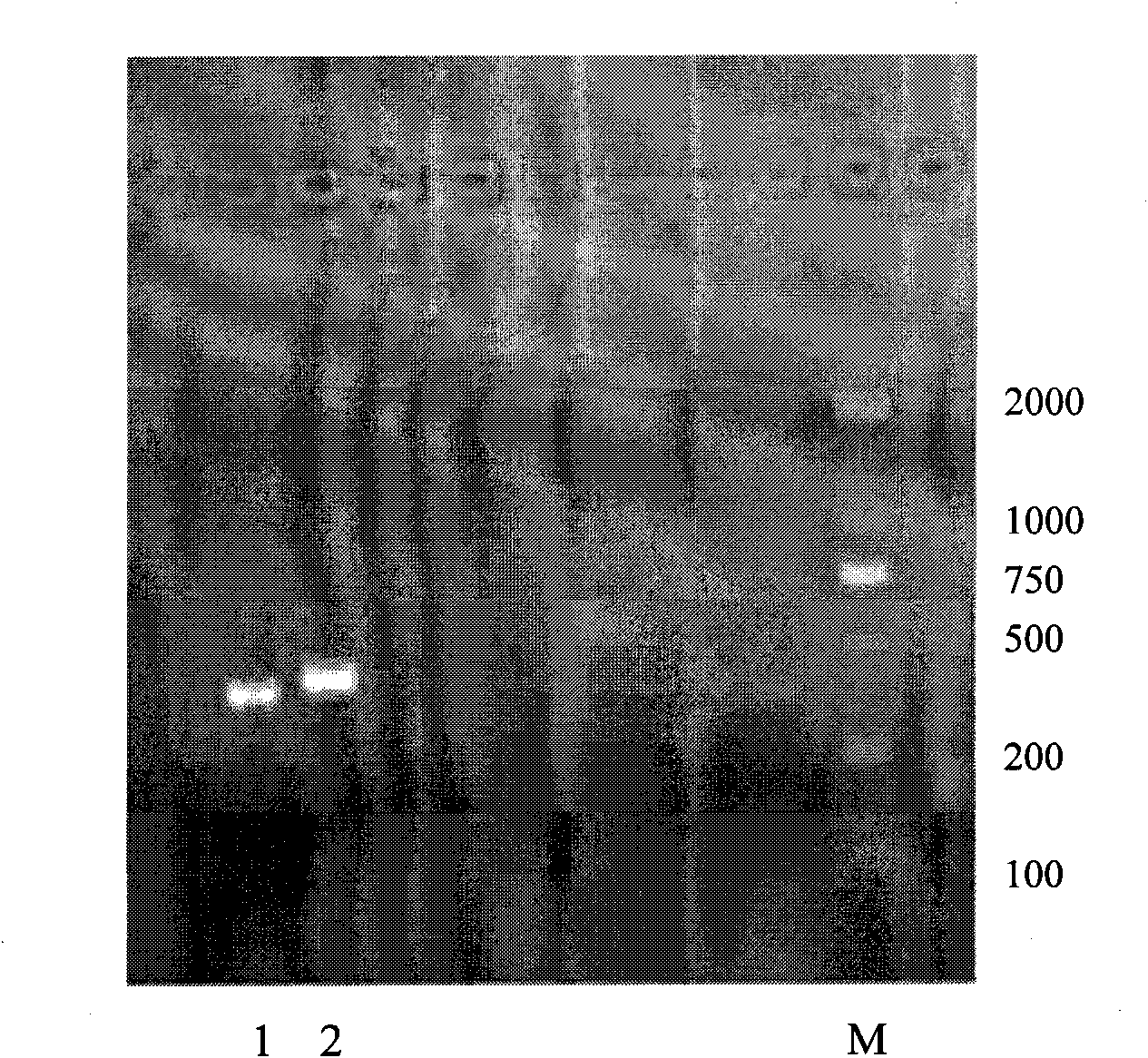
[0058] The result is as figure 1 As shown, band 2 represents kit exon 11, and its PCR product is 438bp.
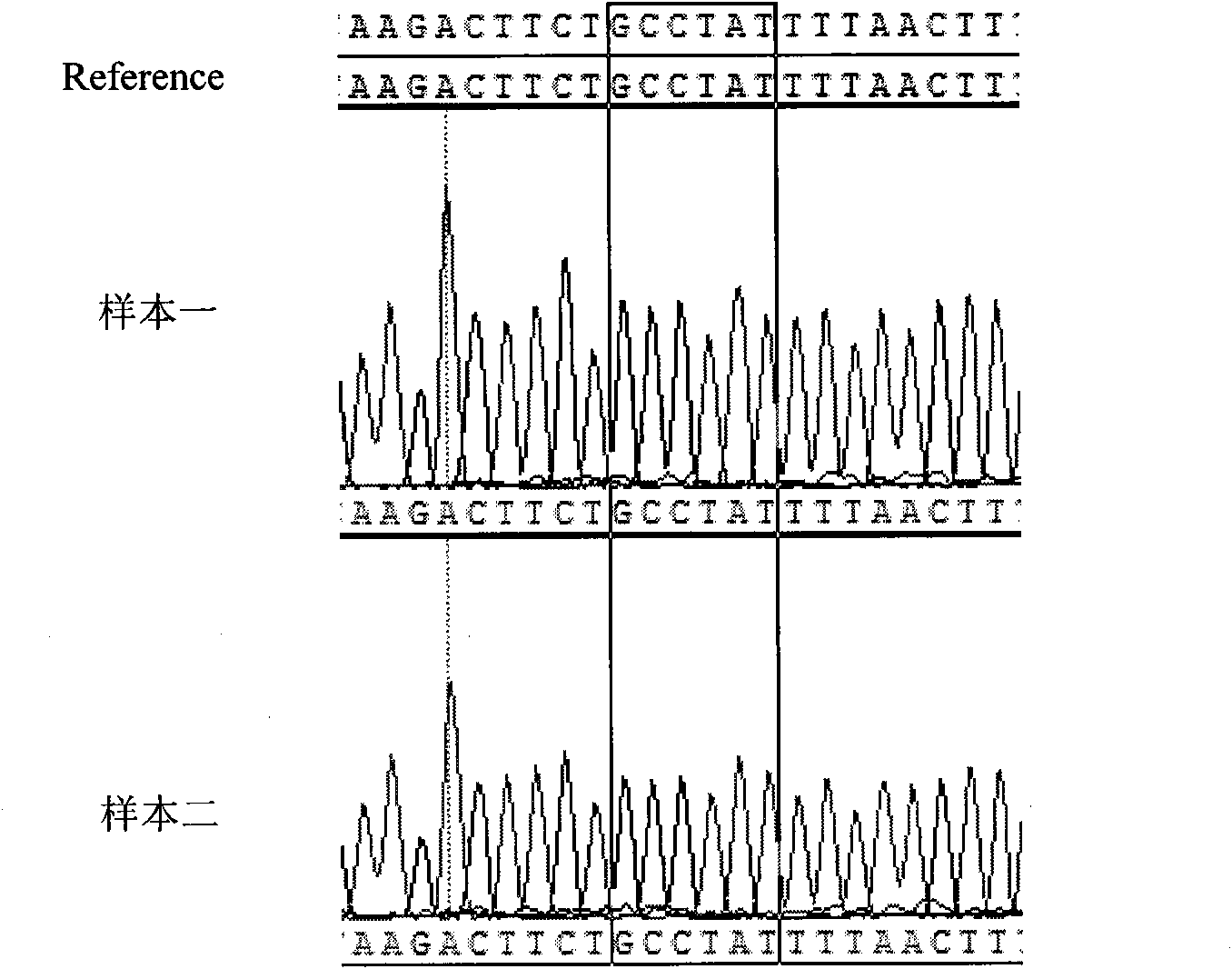
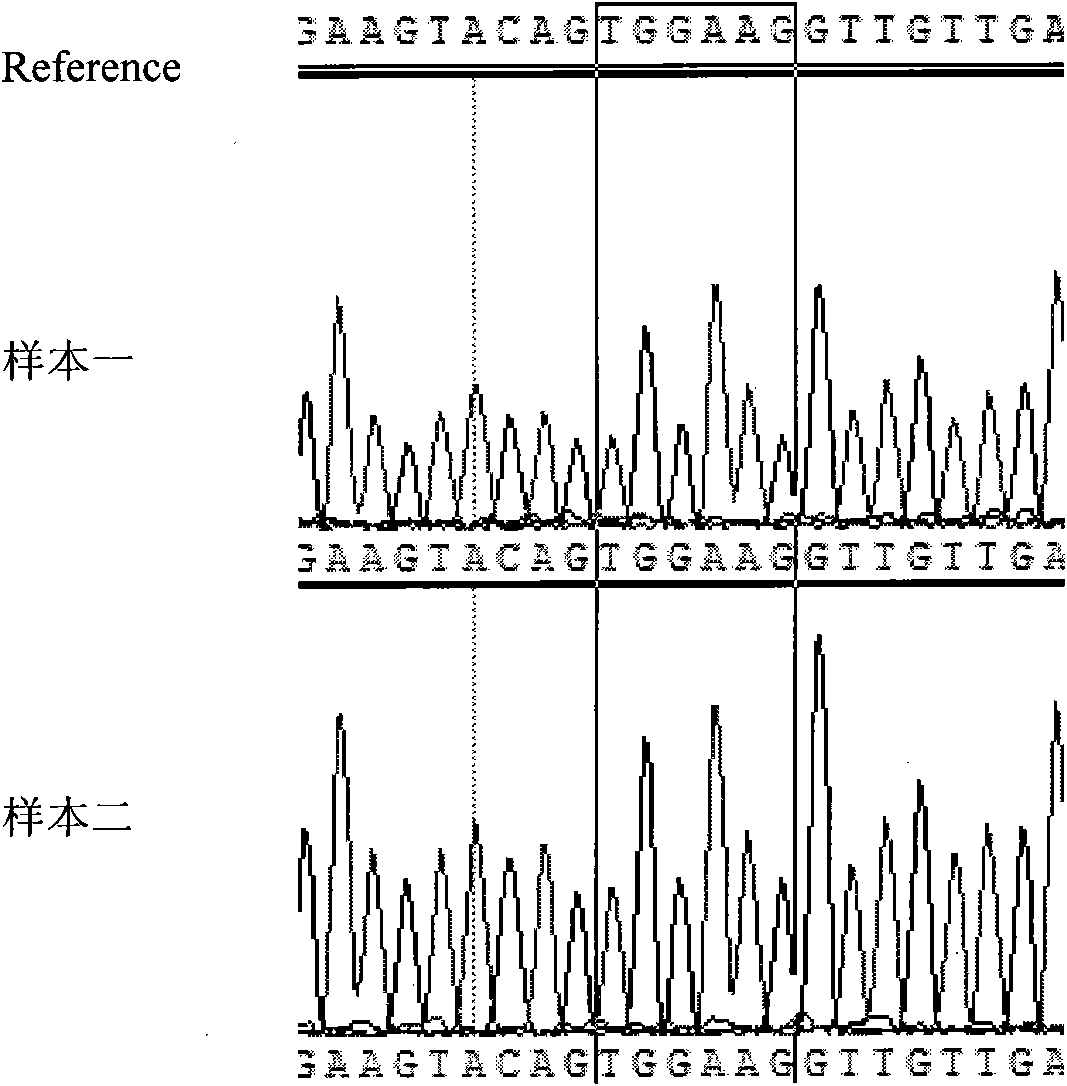
[0059] 4. PCR amplification product sequencing, the specific steps are ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com