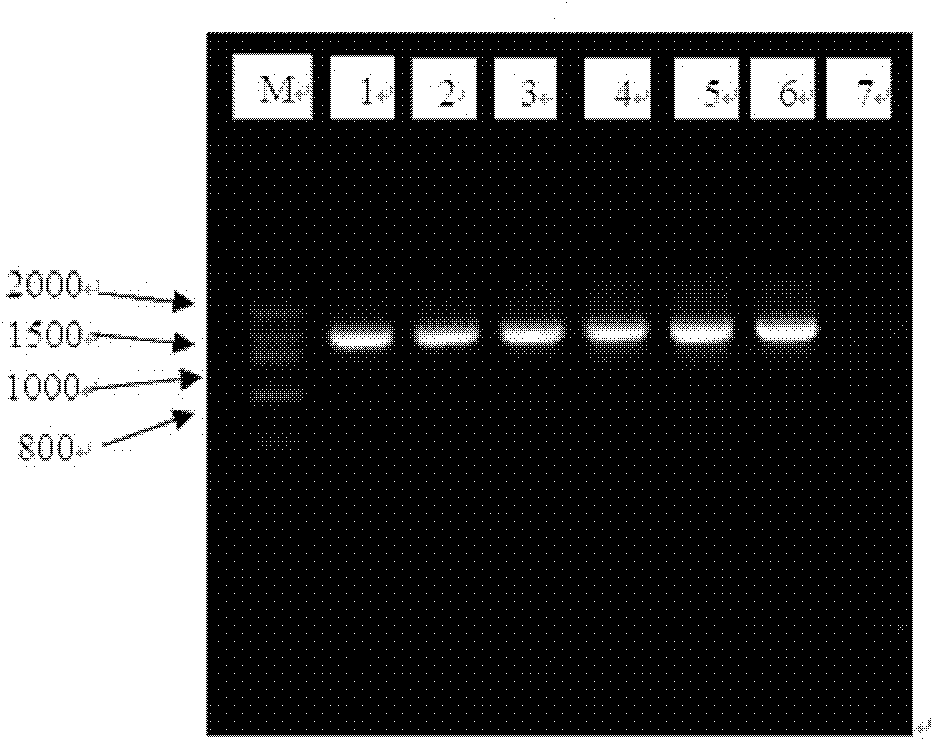
Simple extraction method for DNAs of microbes in river environment sample
A technology of microorganisms and microbial cells, which is applied in the fields of environmental science and bioengineering, can solve the problems of small amount of DNA, many steps of microbial DNA extraction, long operation time, etc., and achieve a strong practical effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Extraction of microbial DNA in Tianjin Haihe environmental samples
[0044] 1. Extraction of microbial DNA in sediment samples
[0045] (1) Lysis of microbial cells in sediment samples:
[0046] Weigh 1 g of bottom sludge into a 10 mL sterile centrifuge tube, add 2 mL of DNA Extraction Buffer, vortex for 10 minutes, add 1 mL of SDS with a mass concentration of 20%, and vortex thoroughly. Place the centrifuge tube in liquid nitrogen for 30 seconds, then place it in a 65°C water bath for 20 minutes; repeat this operation 1-2 times. The samples were taken out and centrifuged at 8000rpm for 15 minutes. Transfer the supernatant to another 10mL sterile centrifuge tube for later use.
[0047] Add 2mL DNA Extraction Buffer and 0.5ml 50mg / mL lysozyme to the precipitate, and vortex until mixed. Water bath at 37°C for 2 hours, add 1 mL of SDS with a mass concentration of 20%, bathe in water at 65°C for 30 minutes, gently shake the centrifuge tube 2-3 times in the mi...
Embodiment 2
[0063] Embodiment 2: Extraction of microbial DNA in different river environment samples
[0064] 1. Extraction of microbial DNA from sediment samples of the Hunhe River in Liaoning
[0065] (1) Lysis of microbial cells in sediment samples:
[0066] Weigh 1 g of bottom sludge into a 10 mL sterile centrifuge tube, add 2 mL of DNA Extraction Buffer, vortex for 10 minutes, add 1 mL of SDS with a mass concentration of 20%, and vortex thoroughly. Place the centrifuge tube in liquid nitrogen for 30 seconds, then place it in a 65°C water bath for 20 minutes, and repeat this operation 1-2 times. The samples were taken out and centrifuged at 8000rpm for 15 minutes. Transfer the supernatant to another 10mL sterile centrifuge tube for later use. Add 2mL DNA Extraction Buffer and 0.5ml 50mg / mL lysozyme to the precipitate, and vortex until mixed. Water bath at 37°C for 2 hours, add 1 mL of SDS with a mass concentration of 20%, bathe in water at 65°C for 30 minutes, gently shake the cent...
Embodiment 3
[0082] Embodiment 3: the extraction of other types of microbial DNA
[0083] (1) Soil microbial cell lysis in Tianjin area:
[0084] Weigh 1 g of soil into a 10 mL sterile centrifuge tube, add 2 mL of DNA Extraction Buffer, vortex for 10 minutes, add 1 mL of SDS with a mass concentration of 20%, and vortex thoroughly. Place the centrifuge tube in liquid nitrogen for 30 seconds, then place it in a 65°C water bath for 20 minutes, and repeat this operation 1-2 times. The samples were taken out and centrifuged at 8000rpm for 15 minutes. Transfer the supernatant to another 10mL sterile centrifuge tube for later use. Add 2mL DNA Extraction Buffer and 0.5ml 50mg / mL lysozyme to the precipitate, and vortex until mixed. Water bath at 37°C for 2 hours, add 1 mL of SDS with a mass concentration of 20%, bathe in water at 65°C for 30 minutes, gently shake the centrifuge tube 2-3 times in the middle, and centrifuge at 8000rpm for 15 minutes. Mix the supernatant with the previous supernat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com