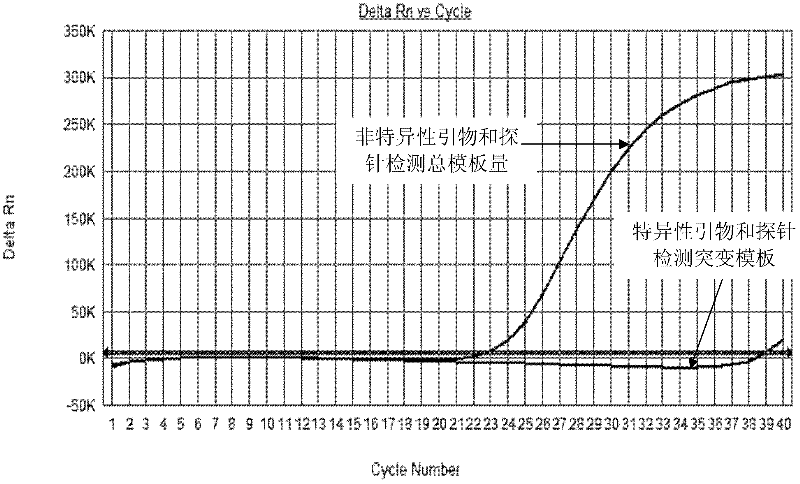
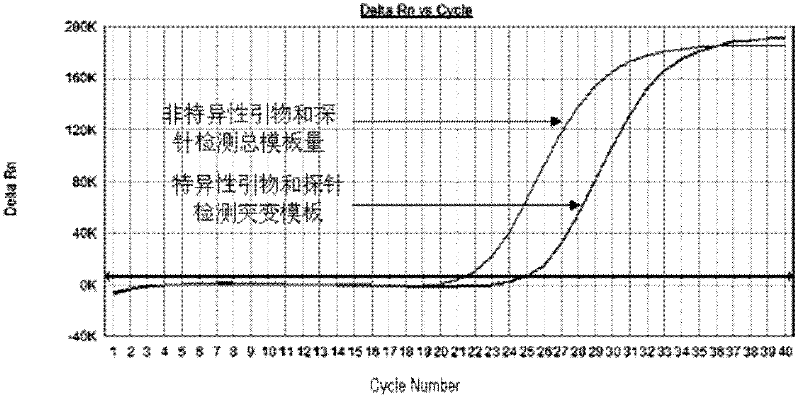
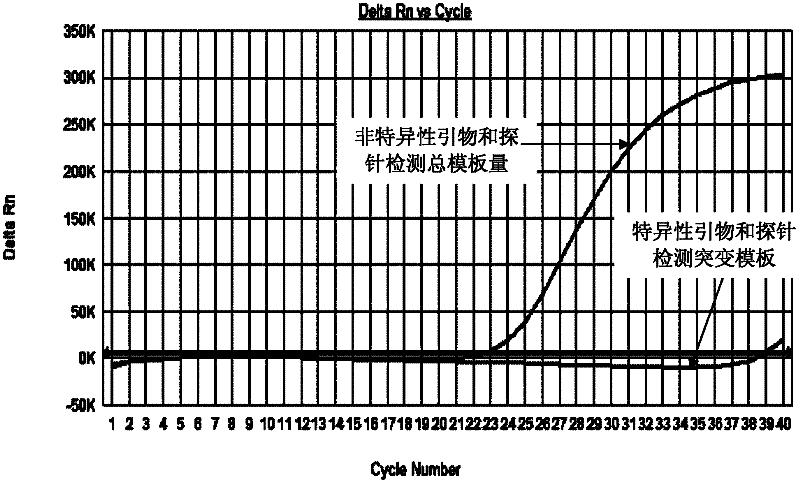
Primers and probes for detecting mutation of cancer gene BRAFV600E
An oncogene and probe technology, which is applied in the field of multiple primers and probes for the detection of oncogene BRAFV600E mutation, to achieve strong specificity, convenient quality control, and avoid false negative results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment example 1
[0035] (1) The mutation-specific upstream primer BRAFFE-1 sequence is:
[0036] 5'-GTGATTTTGGTCTAGCTACTGA-3'.
[0037] (2) The sequence of the mutant non-specific upstream primer BRAFFV-1 is:
[0038] 5-AGTAAAAATAGGTGATTTTGGTCTAG-3'.
[0039] (3) The general downstream primer BRAFR-1 sequence is:
[0040] 5'-GGCCAAAAATTTAATCAGTGGA-3'.
[0041] (4) Locked nucleic acid probe sequence:
[0042] 5'-FAM-GGT[+C]CCA[+T]CAG[+T]TTG[+A]ACA-BHQ 1-3'.
Embodiment example 2
[0044] (1) The mutation-specific upstream primer BRAFFE-1 sequence is:
[0045] 5'-GTGATTTTGGTCTAGCTACTTA-3'.
[0046] (2) The sequence of the mutant non-specific upstream primer BRAFFV-1 is:
[0047] 5-AGTAAAAATAGGTGATTTTGGTCTAG-3'.
[0048](3) The general downstream primer BRAFR-1 sequence is:
[0049] 5'-GGCCAAAAATTTAATCAGTGGA-3'.
[0050] (4) Locked nucleic acid probe sequence:
[0051] 5'-FAM-GGT[+C]CCA[+T]CAG[+T]TTG[+A]ACA-BHQ1-3'.
Embodiment example 3
[0053] (1) The mutation-specific upstream primer BRAFFE-1 sequence is:
[0054] 5'-AAAAATAGGTGATTTTGGTCTAGCTACATA-3'.
[0055] (2) The sequence of the mutant non-specific upstream primer BRAFFV-1 is:
[0056] 5'-AAAAATAGGTGATTTTGGTCTAGCTACA-3'.
[0057] (3) The general downstream primer BRAFR-1 sequence is:
[0058] 5'-TAGTTGAGACCTTCAATGACTTTCTTAGTAA-3'.
[0059] (4) TaqMan probe sequence:
[0060] 5'-FAM-ACAACTGTTCAAACTGATGGGACC-BHQ1-3'.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com