Real-time fluorescence quantitative PCR method for detecting sugarcane ratoon stunning disease
A real-time fluorescence quantitative and dwarf disease technology, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microbial measurement/inspection, can solve the problems of laborious, inability to quantify the number of pathogenic bacteria, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Extraction of pathogenic bacteria of dwarfism in perennial roots by CTAB method Leifsonia xyli subsp .xyli (Preserved in the Sugarcane Comprehensive Research Institute of Fujian Agriculture and Forestry University. The method for isolating the pathogenic bacteria of sugarcane perennial dwarf disease is provided by Y.-B. Pan). Using the DNA template, qualitative PCR amplification was performed using the above primers. Eppendorf Mastercycler EP PCR Thermal Cycler Model 5333 Gene Amplifier. 25 μL PCR reaction system composition: 10×PCR Buffer (Mg 2+ ) 2.5 μL, dNTP (2.5 mmol / L) 2 μL, forward and reverse primers (10 μmol / L) 1 μL each, ExTaq enzyme (5U / μL) 0.125 μL, template DNA (100 ng / μL) 1 μL, add Sterilize double distilled water to bring the total volume to 17.375. PCR amplification conditions: 95°C for 5 min; 35 cycles of 95°C for 30 S, 56°C for 30 S, and 72°C for 30 S; finally extend at 72°C for 7 min and store at 4°C for later use. After the reaction, the obtained...
Embodiment 2
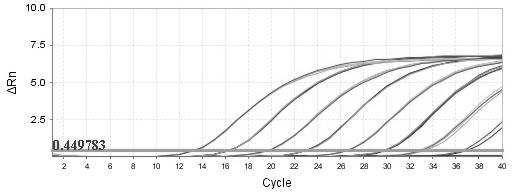
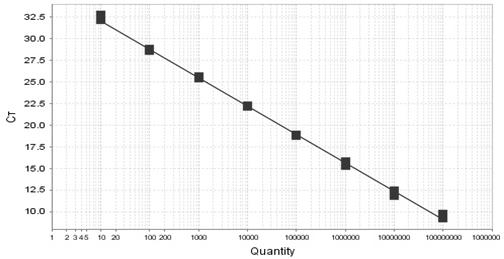
[0057] Sugarcane juice DNA was extracted from sugarcane in the Ninth National Sugarcane Regional Experiment in Fujian Agriculture and Forestry University in 2011. The 11 different varieties of sugarcane are as follows: YA05-179, Liucheng LC1, YG35, YG34, ROC20, FN36, YZ05-5, ROC22, RC16, Y06-407, LC03-1137. According to the traditional nucleic acid extraction method CTAB method, the purified genomic DNA was used as a template, and the 7500 real-time fluorescent quantitative PCR instrument of ABI Company was used to perform PCR amplification with the specific primer pair and TaqMan probe of the present invention. Reaction system includes: TaqMan Universal Master Mix (2×) 12.5 μL; forward and reverse primers (10 μmol / L) 0.75 μL each; TaqMan probe (10 μmol / L) 0.40 μL; DNA template 1.0 μL; , each sample was repeated 3 times. Reaction conditions: 50 ℃, 2 min, pre-denaturation 95 ℃, 10 min; 95 ℃, 15 s; 60 ℃, annealing extension, 1 min, 40 cycles, single-point fluorescence detection...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com