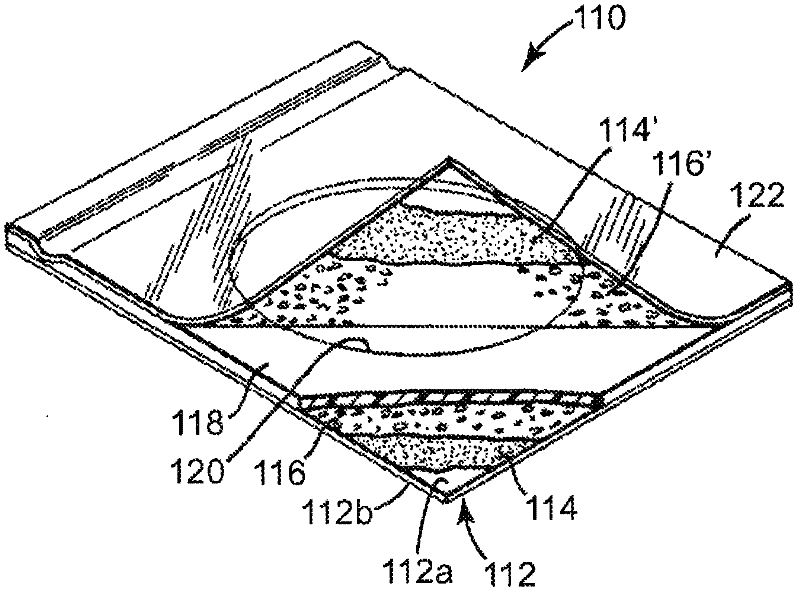
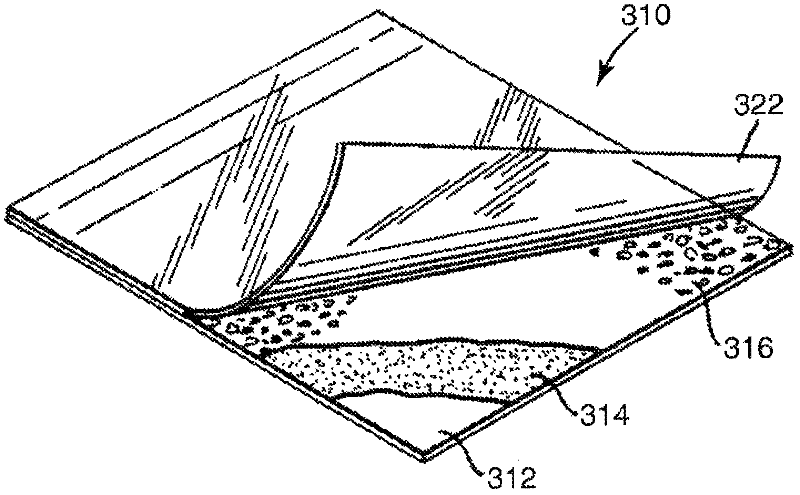
Methods and articles for detecting deoxyribonuclease activity
A deoxyribonucleic acid and enzyme activity technology, applied in the field of detection of deoxyribonuclease activity and products, can solve the problems of laborious cultivation methods and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0144] Detection of Growth and DNase Activity in PETRIFILM Aerobic Counting Plates
[0145] Phosphate buffered saline (PBS) was obtained as a 1OX concentrate (OmniPur 6505; EMD Chemicals; Gibbstown, NJ). DNA (sodium salt, from salmon testes) and methyl green dye (zinc chloride salt, Aldrich 198080-10G) were obtained from Sigma-Aldrich (St. Louis, MO). Lambda-carrageenan (viscarin GP type 109F) was obtained from FMC BioPolymer (Philadelphia, PA). 3M PETRIFILM Aerobic Counting Plates and PETRIFILM Plate Reader (PPR) were obtained from 3M Company (St. Paul, MN). Staphylococcus aureus ATCC 25923 (DNase positive, methicillin sensitive) was obtained from the American Type Culture Collection (Manassas, VA). S. epidermidis strain 472 (DNase negative) and S. aureus strain 565 (DNase positive, methicillin resistant) were obtained from clinical isolates.
[0146] A solution of DNA-PBS solution was prepared by adding 2.0 mg / mL DNA and 0.1 mg / mL lambda carrageenan to IX (10 mM) PBS. ...
example 2
[0150] Analysis of Petrifilm plate images
[0151] PETRIFILM Aerobic Count Plates were prepared as described in Example 1. Plates were inoculated with S. aureus Staphylococcus ATCC 25923 (a DNase producing microorganism). After seeding, the hydrogel in the plate contained 20 μg / mL methyl green. Plates were imaged using a PETRIFILM plate reader as described in Example 1.
[0152] Red, green and blue images were exported to IMAGE-PRO Plus version 6.3.0.512 software. Illustrations of grayscale images of the board when illuminated with green or red LEDs are in Figure 7a and 8a displayed in . The graticules of the plate (which are clearly visible in the green illuminated image but not in the red illuminated image) have been omitted from the figure.
[0153] Figure 7a The hydrogel 782 in the inoculated portion of the plate is shown. Several bacterial colonies 784 are also shown. Using the line profile tool of Image Pro Plus software, the region of interest from pixel x,...
example 3
[0160] Early Detection of DNase-producing Microorganisms
[0161] PBS-DNA solutions with (50 μg / mL) and without methyl green were prepared as described in Example 1. Use a final methyl green concentration of 20 µg / mL in this experiment in all plates containing methyl green.
[0162] A suspension of washed bacterial cells (S. aureus ATCC strain 25923) was prepared as described in Example 1. The suspension was diluted and inoculated into Petrifilm aerobic count plates as described in Example 1 .
[0163] The inoculated plate was placed on top of a metal heat block set at 37°C. A NIKON E8400 digital camera (Nikon, Inc., Melvelle, NY) was placed on top of the plate. The camera was connected to a computer via IMAGE-PRO Plus version 6.3.0.512 software, which was programmed to take digital photographs of each plate every 30 minutes for 24 consecutive hours. Place an insulated lid over the camera and heat block apparatus to maintain the temperature of the plate at 37 °C. The pr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com