SYBR-Green fluorescent quantitative RT-PCR (reverse transcription-polymerase chain reaction) detection method for Microcystis aeruginosa phycophages in water environment
A microcystis green algae phagocytosis and a microcystis green algae phagocytosis technology are applied in the field of molecular biology and can solve the problem that there are not many methods for real-time quantitative changes of algae phages.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
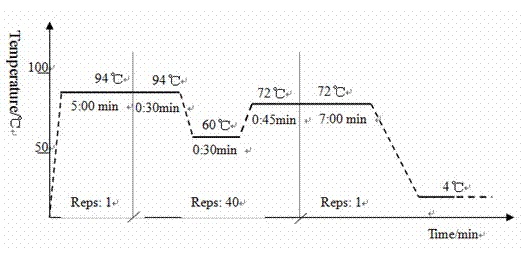
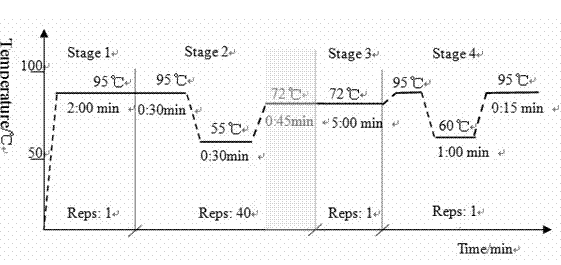
[0035] The experimental methods not indicating specific conditions in the following methods are usually according to conventional conditions such as the molecular cloning of Sambrook et al.: Laboratory Manual (New York: Cold Spring Harbor Laboratory Press, 1989) or the genetic engineering experimental technique of Peng Xiuling et al. (Science Technical Press), or as recommended by the manufacturer. The gene sequences of cyanophages were obtained from the Molecular Biology Database (NCBI).
[0036] 1. PCR primer design
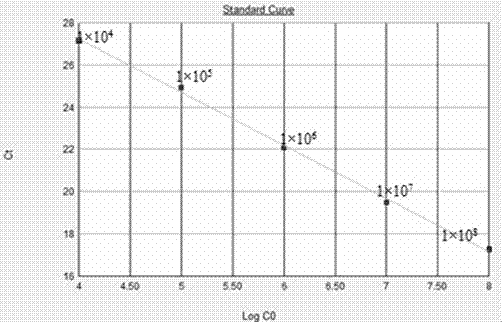
[0037] According to the literature and combined with the BALST sequence alignment results in Genebank, refer to the tail sheath protein coding gene (g91) of Microcystis aeruginosa cyanophage Ma-LMM01, NCBI sequence number is AB242261, and use Primer Premier 5.0 and Oligo6 software to design primers . The quantitative PCR external reference standard primers SH-WF and SH-WR, quantitative primers SH-RTF and SH-RTF (Table 1) were designed, and the designed pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com