Probe set for identification of nucleotide mutation, and method for identification of nucleotide mutation
A technology of probe group and nucleotide, which is applied in the field of probe group for nucleotide mutation identification and nucleotide mutation identification, which can solve insufficient problems and achieve the effect of simple research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
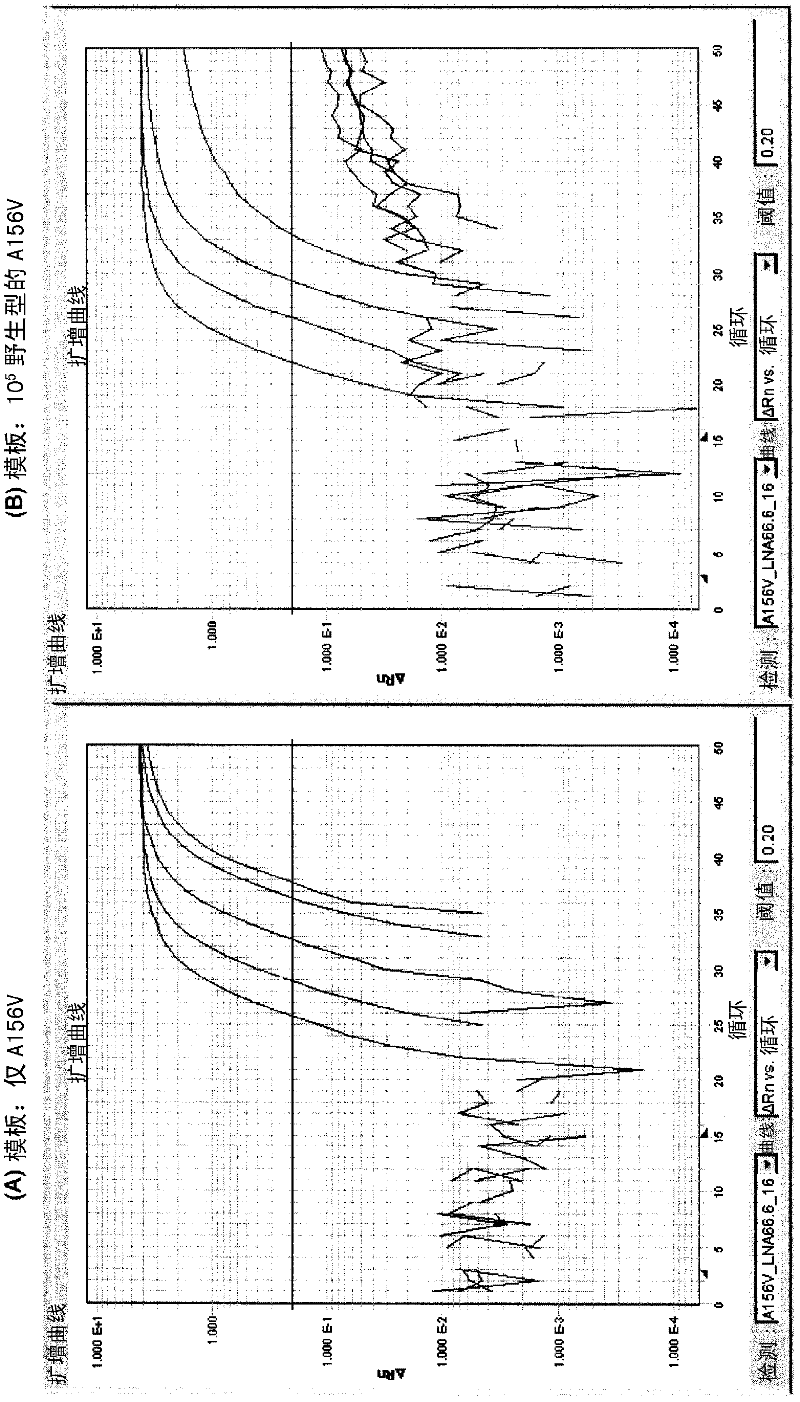
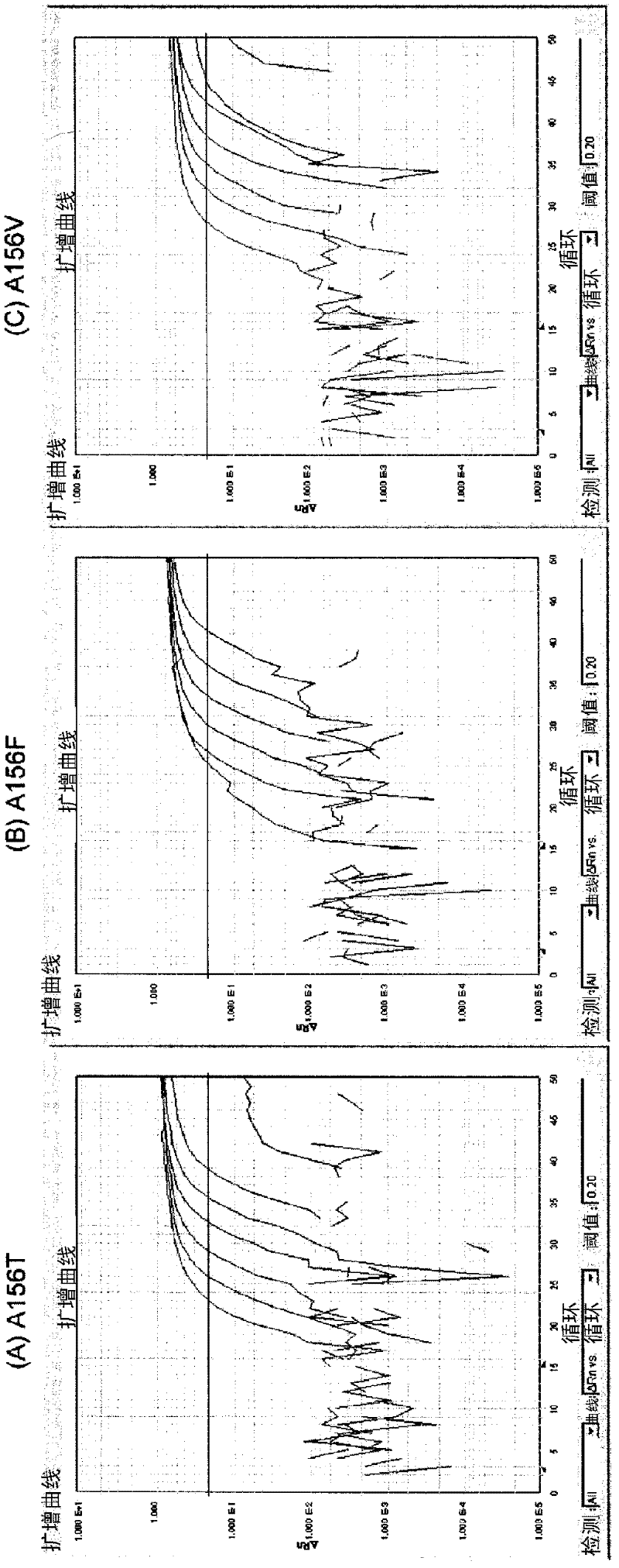
[0243] Amino acid at position 156 of HCV-1b NS3 protease is Ala in the wild type, and as a result of nucleotide substitution, it is replaced with Val, Phe, Thr or Ser, etc. in the mutant virus. Here, real-time PCR was performed using the following fluorescent probes for the specific detection of mutations leading to amino acid substitutions at position 156 to Val, more specifically, the nucleotide encoding the NS3 protease Mutation of C→T substitution at position 467 of the sequence.
[0244]
[0245] In each of the upstream and downstream sides of the nucleotides encoding the 156-position amino acid, the primers were fixed to the HCVNS3 protease in patients enrolled in clinical trials of Teraprevir and in a public database (The Entrez Nucloetide Database) In the consensus region between the regions (positions 289 to 305 (Fw primer) and 487 to 503 (Re primer) of SEQ ID NO: 1.
[0246] Fw primer 5'-TGCACCTGCGGCAGCTC-3' (SEQ ID NO: 3)
[0247] Tm = 69.2°C (at the salt concen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com