Swine pseudo attp site and use of swine pseudo attp site
A promoter and optional technology, applied in the field of bioengineering, can solve problems such as unfavorable cultivation of stable strains, inactivation, destruction of endogenous genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1, the construction of the pCMV-Int vector containing streptomyces phage ΦC31 integrase gene
[0064] (1) Synthesis of Streptomyces phage φC31 integrase gene (Int gene): Submit the sequence shown in SEQ ID NO: 5 to Shanghai Jierui Biotechnology Co., Ltd. for Int gene synthesis, and construct the synthesized Int gene in pGH vector On, get the pGH-Int plasmid.
[0065] (2) Enzyme digestion of pGH-Int plasmid: Take 20 microliters (400-500ng / ul) of pGH-Int plasmid for enzyme digestion. The enzyme digestion system is:
[0066] pGH-Int plasmid
20 microliters
10xNEB buffer2
5 microliters
endonuclease NheI
1 microliter
1 microliter
[0067] 100xBSA
0.5 μl
double distilled water
22.5 µl
total reaction volume
50 µl.
[0068] Place the above enzyme digestion system at 37°C and digest overnight.
[0069] (3) Enzyme digestion of pcDNA3.1 plasmid: take 10 mic...
Embodiment 2
[0076] Embodiment 2, the construction of pattB-CAG-Intron-EGFP carrier
[0077](1) Synthesis of attB and Intron gene fragments: Submit the sequence Intron (intron) sequence and the attB sequence shown in SEQ ID NO: 4 to Shanghai Jierui Biotechnology Co., Ltd. for gene synthesis, and construct the synthesized genes separately On the pGH vector, pGH-attB and pGH-Intron plasmids were obtained, respectively. Among them, the NCBI of the used intron is AF369966.1, and the full length of the sequence is
[0078] CGAATCCCGGCCGGGAACGGTGCATTGGAACGCGGATTCCCCGTGCCAAGAGTGACGTAAGTACCGCCTATAGAGTCTATAGGCCCACAAAAAATGCTTTCTTCTTTTAATATACTTTTTTGTTTATCTTATTTCTAATACTTTCCCTAATCTCTTTCTTTCAGGGCAATAATGATACAATGTATCATGCCTCTTTGCACCATTCTAAAGAATAACAGTGATAATTTCTGGGTTAAGGCAATAGCAATATTTCTGCATATAAATATTTCTGCATATAAATTGTAACTGATGTAAGAGGTTTCATATTGCTAATAGCAGCTACAATCCAGCTACCATTCTGCTTTTATTTTATGGTTGGGATAAGGCTGGATTATTCTGAGTCCAAGCTAGGCCCTTTTGCTAATCATGTTCATACCTCTTATCTTCCTCCCACAGCTCCTGGGCAACGTGCTGGTCTGTGTGCTGGCCCATCACTTTGGC...
Embodiment 3
[0084] Embodiment 3, cell transfection and screening
[0085] The porcine kidney epithelial cell line PK15 was inoculated into a 6-well plate at a density of 30%, and the cells were cultured until the confluence was 70-80%, and transfection was started.
[0086] (1) Transfection: The pCMV-Int plasmid obtained in Example 1 and the pattB-CAG-Intron-EGFP plasmid obtained in Example 2 were mixed in different ratios, and the liposome LF2000 kit was used to follow the manufacturer's instructions. Instructions for transfecting PK15 cells.
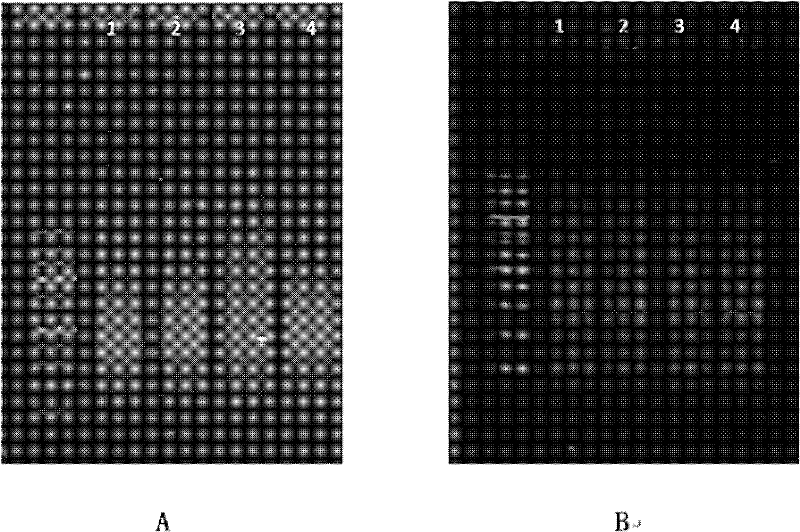
[0087] (2) Exploration of the effect of two plasmid ratios: with the quality of the pattB-CAG-Intron-EGFP plasmid constant at 1 microgram, by selecting the quality of the pCMV-Int plasmid as 1 microgram, 2 micrograms, 3 micrograms, 4 micrograms, and 5 micrograms, The pattB-CAG-Intron-EGFP: pCMV-Int was transfected into PK15 cells according to different ratios of 1:1, 1:2, 1:3, 1:4, and 1:5. The best mixing ratio, the result is that the fluoresce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com