Sequencing method for increasing reading length
A sequencing and read length technology, applied in the field of genetic engineering, can solve the problems of short read length and low reading accuracy of nucleotide sequence information in ligation sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0045]In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments.
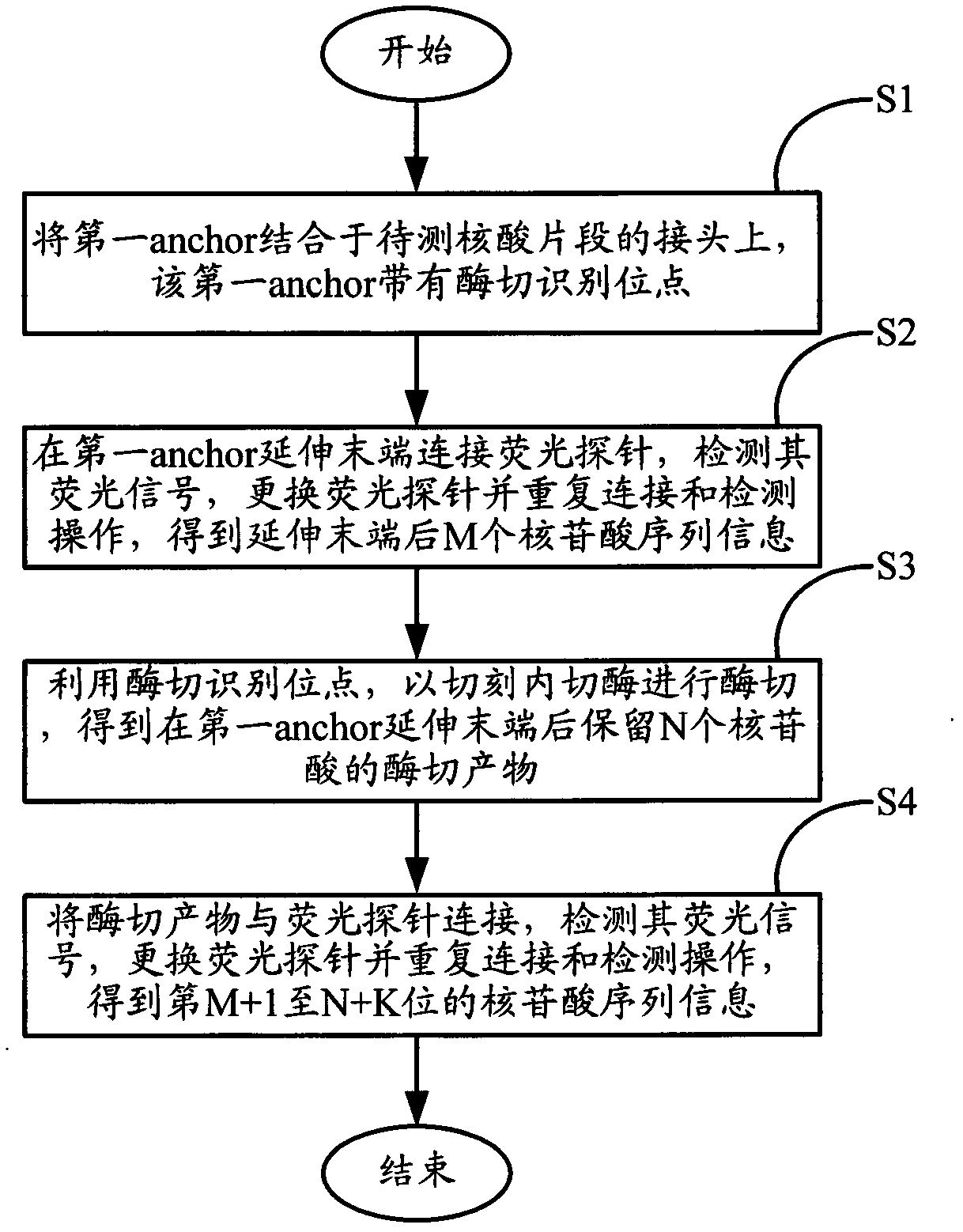
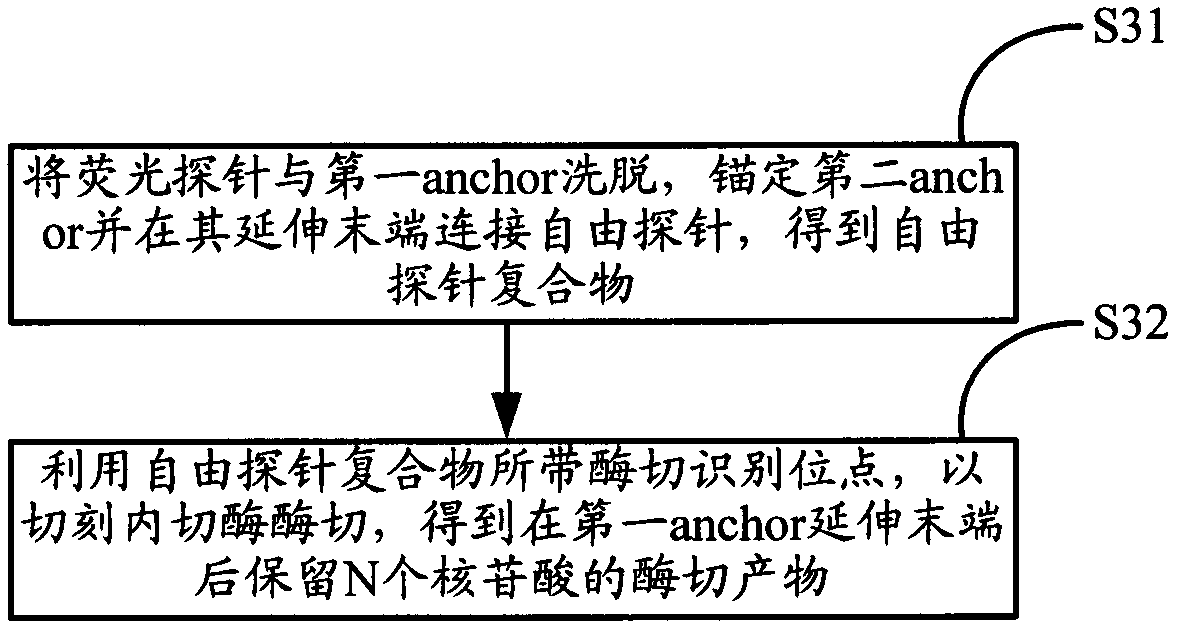
[0046] The present invention provides a sequencing method for increasing the read length, which includes the following steps: firstly, according to the steps of the conventional ligation sequencing method, using the anchor primer anchor and different types of fluorescent probes used for detection, repeatedly hybridize the anchor anchor, connect the fluorescent probe The steps of needle, collecting fluorescent signal, and eluting anchor and fluorescent probe are to read the nucleotide sequence information at the M position after the extended end of the anchor in the nucleic acid fragment to be tested according to the type of the connected fluorescent probe. Wherein, the anchor has a nicking endonuclease recognition site; the extended end of the anchor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com