Method for gathering and separating endogenous nuclear receptors and deoxyribonucleic acid (DNA) binding sequence special for same
A DNA sequence and binding sequence technology, applied in the field of enrichment and separation of endogenous nuclear receptors and their special DNA binding sequences, can solve the problems of limited antibody types, limited separation and identification, low expression level of NR and its co-regulatory proteins, etc. Achieve the effect of solving detection problems and wide application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1. Acquisition of dedicated DNA-binding sequences for enrichment and isolation of endogenous nuclear receptors and their complexes
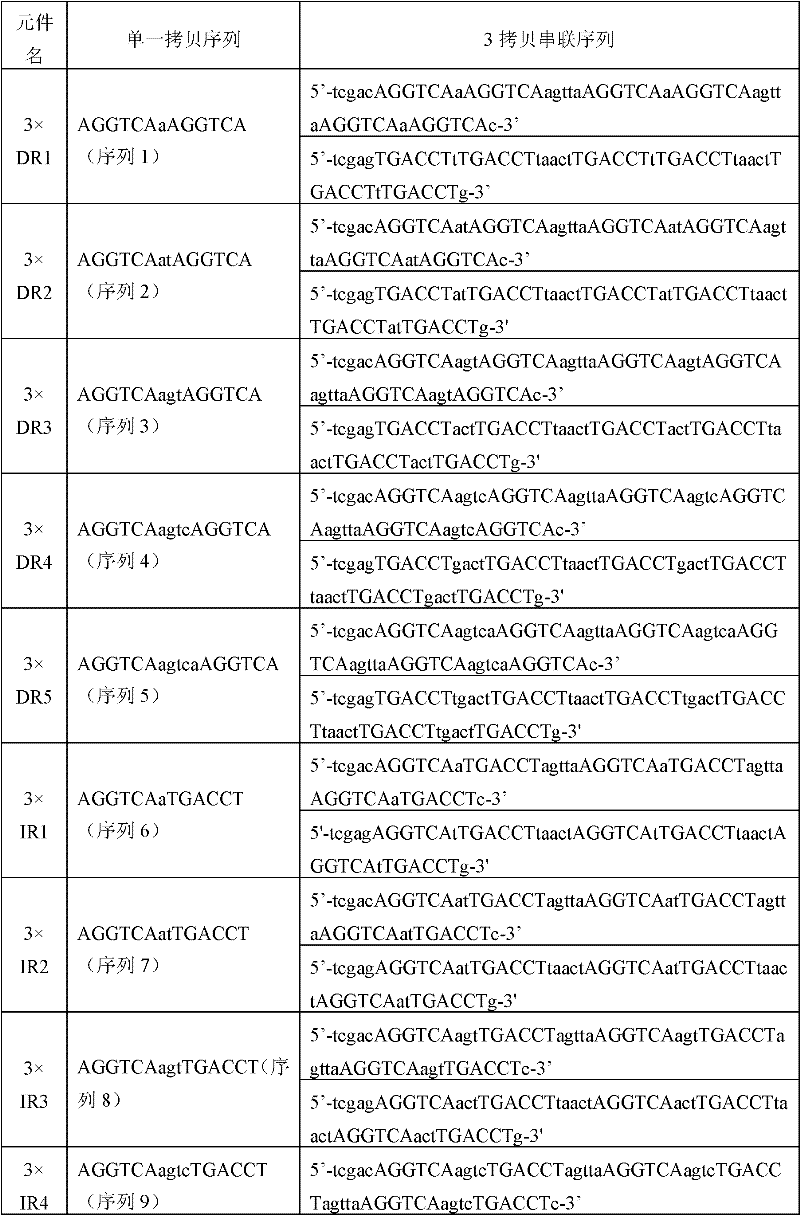
[0030]According to the specific combination of endogenous nuclear receptor (NR) and hormone response element (hormone response element, HRE), and HRE usually contains two conserved half-sites (5'-AGGTCA-3'), the present invention designs A variety of DNA sequences containing conserved half-site elements in which two conserved half-sites are arranged either as direct repeats (DR) or inverted repeats (IR) and where the two half-sites Spots consist of 0-6bp bases. These DNA sequences can form tandem multi-copy HRE sequences through techniques such as in vitro ligation. A single copy of HRE contains two conserved half-sites, and a 3-5bp linker sequence (Linker) is connected in tandem between each single copy of HRE; Table 1 shows the tandem 3 Copy the DNA binding sequence of HRE, in which the sequence 1-17 in the sequence table is a s...
Embodiment 2
[0057] Example 2. Enrichment and separation of endogenous nuclear receptors in mouse liver cells
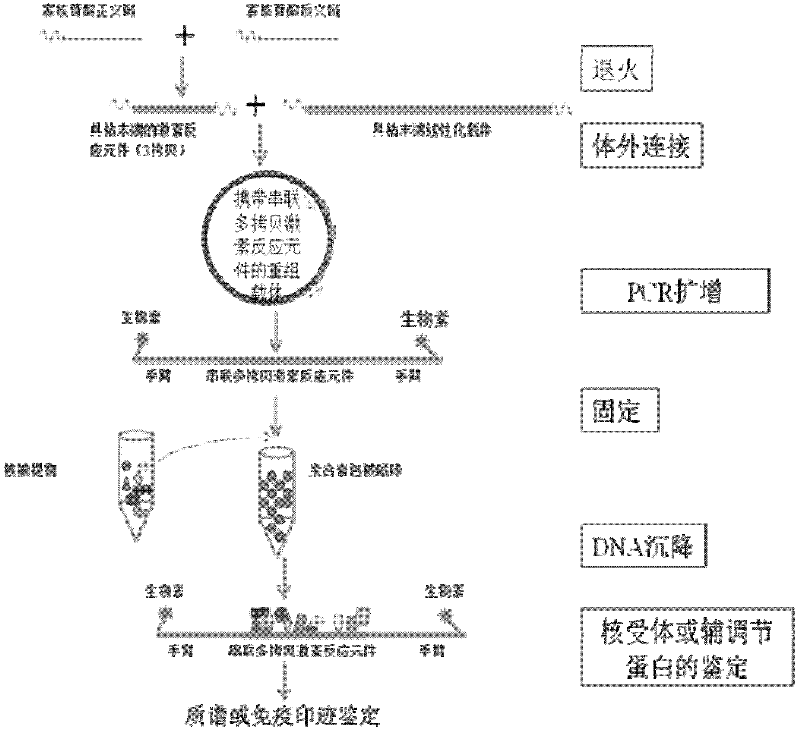
[0058] Taking 12×DR1 as an example, the nuclear receptors in mouse liver cells were enriched and isolated, and identified by mass spectrometry. The enrichment separation referred to in the present invention refers to the separation of nuclear receptors and their complexes from complex nucleoproteins. Using the method of the present invention to enrich and isolate endogenous nuclear receptors in mouse liver cells, such as figure 1 As shown, it specifically includes the following steps:
[0059] 1. Obtain a high-purity biotin-labeled DNA bait (about 600bp fragment) containing 12×DR1 by the method of Example 1;
[0060] 2. Immobilize biotin-labeled DNA bait to streptavidin-coated magnetic beads by biotin-streptavidin binding On M-280streptavodin (Invitrogen), the specific operation is as follows:
[0061] 1) Take 60 μl of magnetic beads into a clean EP tube, place them on a mag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com