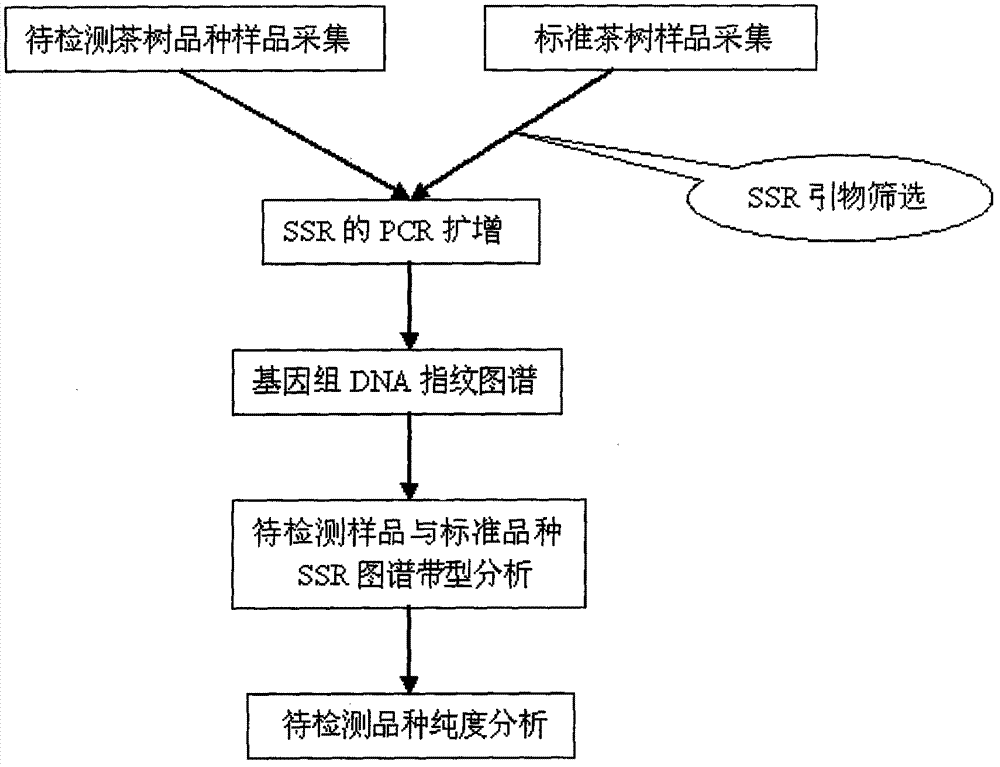
Molecular marking method for identifying tea clonal variety of tea trees
A technology of molecular markers and clones, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as difficulty in effective supervision and arbitration, lack of variety identification methods, and low reliability, etc. The gel and staining are easy to master, the DNA extraction procedure is simplified, and the effect of saving time and reagent costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0091] Embodiment 1, take Foxiang No. 3 as standard tea tree variety identification tea tree variety purity.
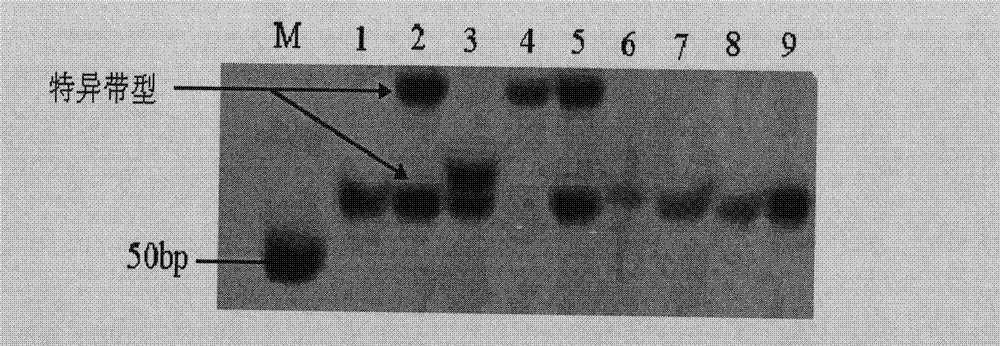
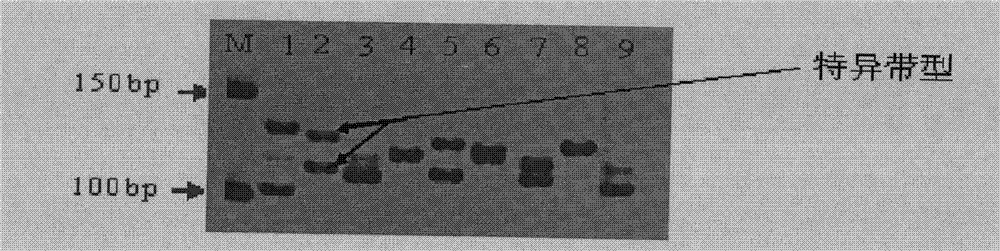
[0092] Before the identification, 27 varieties (including Foxiang 3) from Yunnan Province were screened for SSR markers, and it was found that SSR primers SSR12 and SSR42 could produce specific bands for the genomic DNA of Foxiang 3. Therefore, using primers SSR12 and SSR42 were used to analyze the DNA of samples from Foxiang 3.
[0093] 1. Sample selection
[0094] Select the variety to be tested, Foxiang No. 3, and other 4 varieties to be tested (controls) from the clones of Yunnan tea trees. In order to test the identification effect, they are mixed with tea varieties such as Yunkang No. 10, Foxiang No. 1, and Zijuan. The fresh buds and leaves of one bud and two leaves of the tea tree clone varieties to be tested were collected and stored at -20°C for later use.
[0095] 2. Extraction of Genomic DNA of Tea Tree Varieties
[0096] 1) Put 1.0g of fresh buds and le...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com