Short-day sensitive cell death SSD1 gene and coded protein and application thereof
A cell death and gene encoding technology, applied to the short-day light-sensitive cell death SSD1 gene and its encoded protein and application fields, can solve problems such as complex biological cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1: Arabidopsis EMS mutagenesis and screening
[0021] 1. Chemical mutagenesis treatment
[0022] Arabidopsis wild-type (Col-0) seeds were placed in distilled water and stirred for 30 minutes. After standing at 4°C for 12 hours, the seeds were transferred to 100 mM phosphate buffer (pH 6.5), and 0.2% ( v / v) of ethyl methanesulfonate (EMS), sealed and placed on a shaker in a water bath (25°C) for 12 hours, then rinsed the seeds with distilled water.
[0023] 2. Mutation plant culture
[0024] The rinsed seeds were vernalized at 4°C for 3 days, and sown in artificial soil (Northeast black soil: vermiculite = 1:1, v / v). Grow in the light culture room, the culture temperature is 22±2℃; the light intensity is 80-120μmolm -2 the s -1 ; relative humidity 65%; photoperiod 16 hours light, 8 hours dark. Harvesting mature seeds is M 1 Generation, M 1 M 2 Substitute, dry for later use.
[0025] 3. Screening of Mutants
[0026] Will M 2 After surface disinfectio...
Embodiment 2
[0028] Embodiment 2: Map-based cloning method to isolate SSD1 gene
[0029] 1. Construction of map-based cloning populations
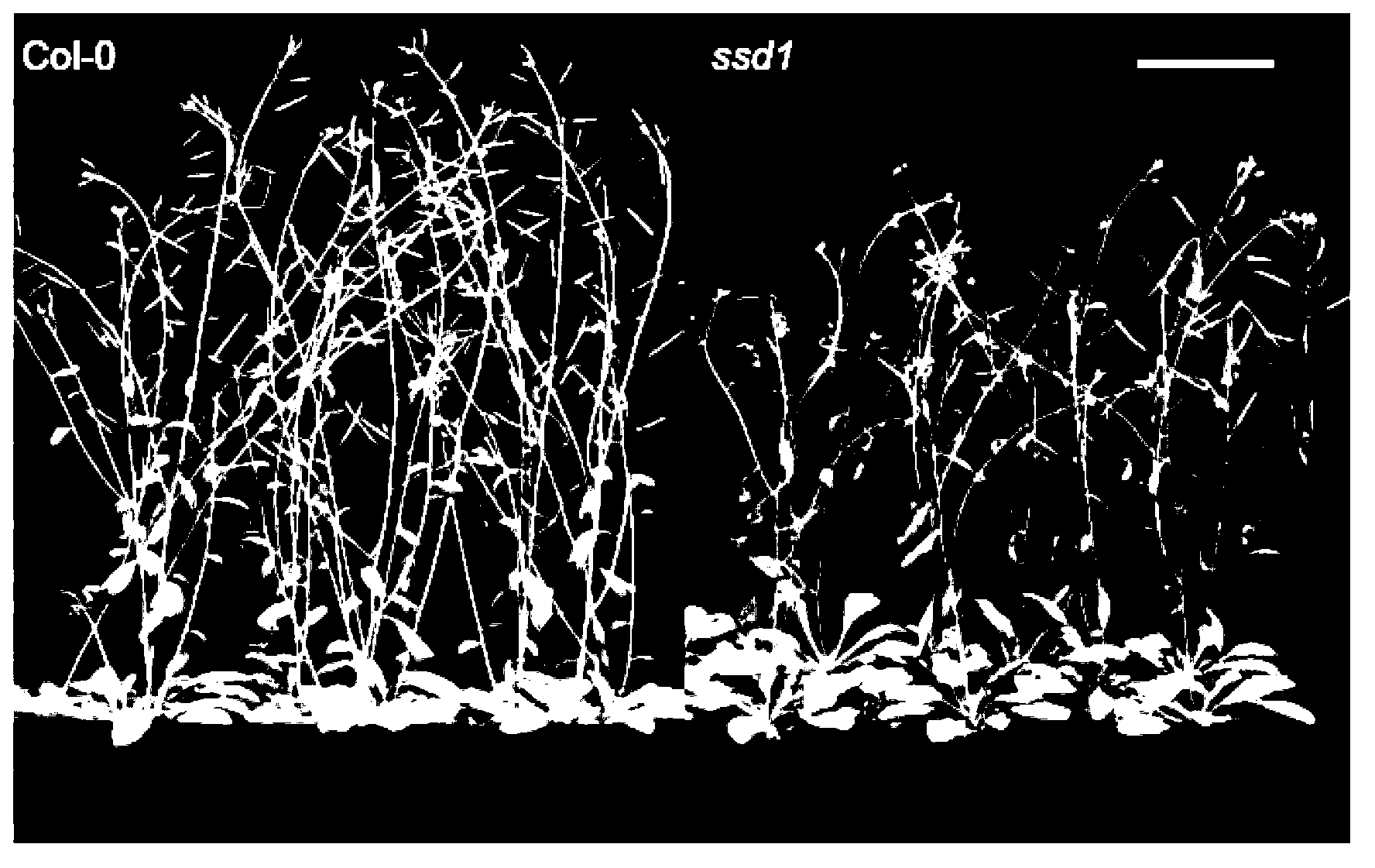
[0030] Crossing the ssd1 mutant (Col-0 background) with another ecotype Landsberg erecta (Ler) gave F 1 Generation heterozygote; F 1 F 2 generation; seeding F 2 Generation seeds were screened for individuals with cell necrosis under short-day conditions, and a row of ssd1 mutants were planted in the same seedling tray as a control. A total of 2500 strains of F 2 Genetic populations are used for map-based cloning of genes.
[0031] 2. Selection and design of molecular markers
[0032] According to the Arabidopsis genome-wide DNA polymorphism database published on TAIR (www.arabidopsis.org), molecular markers were selected: NT7123, EAT, F21M12, NGA63, F16J7-TRB, JV18 / 19, F21J9-83201. According to literature reports, the molecular marker SGCSNP9772 was designed, see Table 1. In addition, based on the alignment of the Ler-sequenced expressed sequen...
Embodiment 3
[0049] Embodiment 3: Genetic complementation experiment of SSD1 gene
[0050] 1. Construction of Complementary Vectors
[0051] The high-fidelity enzyme TransStart FastPfu DNA Polymerase (TransGen) was used to amplify a fragment containing the SSD1 gene from Arabidopsis wild-type Col-0, with a full length of 3175bp. CCCGGG ATGGCGTTGCTGAAGTCTTT-3') and RP(5'-cga GAGCTCAC TTATTGTTAATGGGTTGG-3'). The amplification reaction system is 50μl, of which 5×TransStart FastPfu Buffer: 10μl; 2.5mM dNTP: 5μl; 10mM forward primer FP: 2μl; 10mM reverse primer RP: 2μl; template Col-0 DNA: 0.5-2ng; TransStart FastPfu DNA Polymerase: 1μl; add ddH 2 O to make up to 50 μl. The amplification program was: 95°C pre-denaturation for 2 minutes; 95°C for 20s, 55°C for 20s, 72°C for 2min, 30 cycles; 72°C for 5 minutes.
[0052] Carrier plasmid pROK2 and purified PCR product were digested with SmaI and SacI, and T4 DNA Ligase (TransGen) was used to connect the digested and purified AtSSD1 gene and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com