Nucleic acid detection method applying multiplex PCR array technology
A technology for detecting and detecting primers, which is applied in the biological field and can solve the problems of different lengths, difficulty in setting the annealing temperature program of the mPCR system, and reducing the detection sensitivity of the mPCR.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1 Application of mPCR detection in a single detection object
[0073] Taking the detection of Acinetobacter baumannii (Ab) culture as an example, the present invention relates to the application of the mPCR system. Since the samples used are all known Ab cultures, this example aims to illustrate the detection sensitivity of the method of the present invention and ordinary mPCR without UT sequence. 1. Sample collection.
[0074] Ab clinical isolates were obtained from the local Centers for Disease Control and Prevention (CDC).
[0075] 2. Acquisition of Ab genome information.
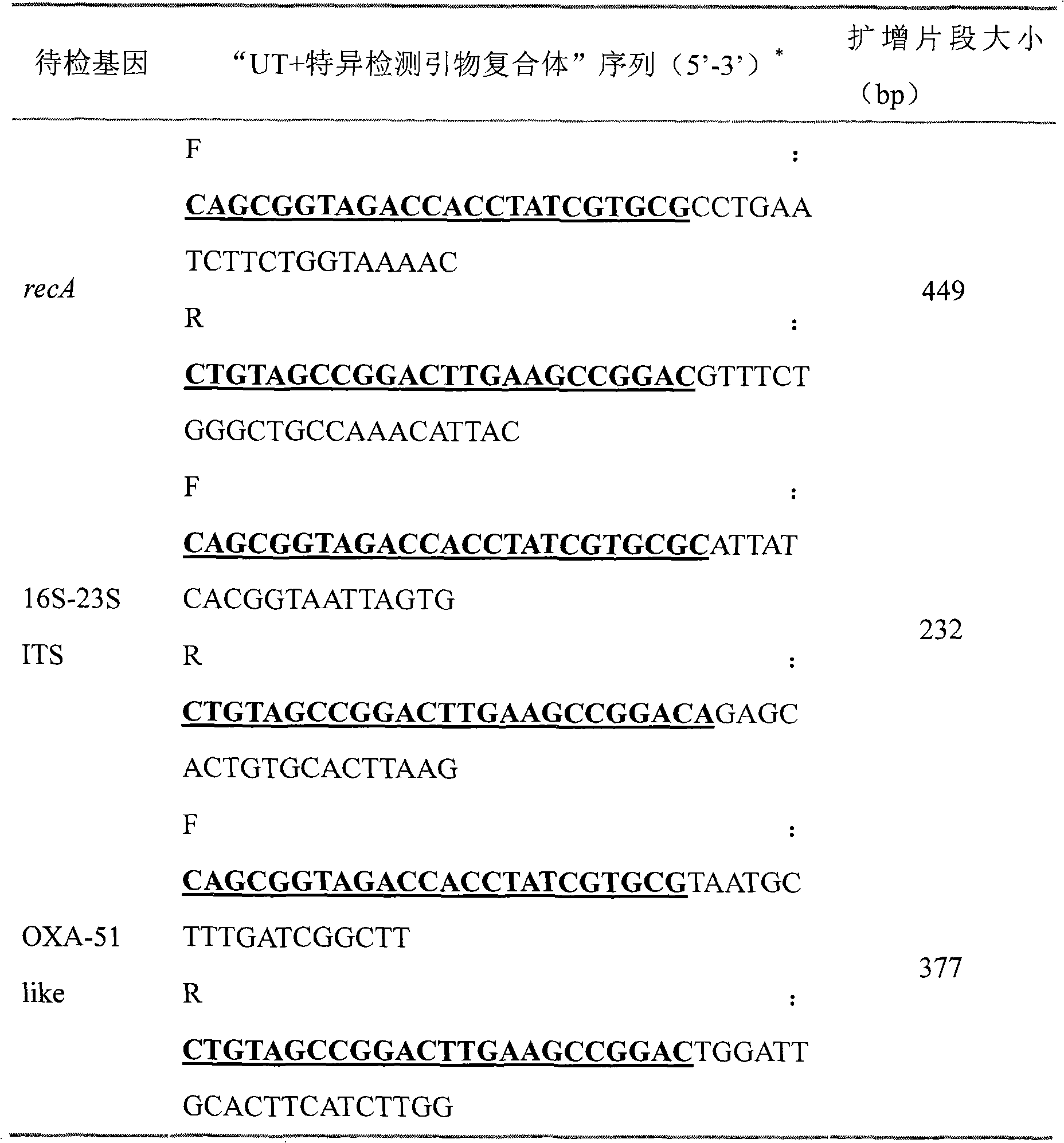
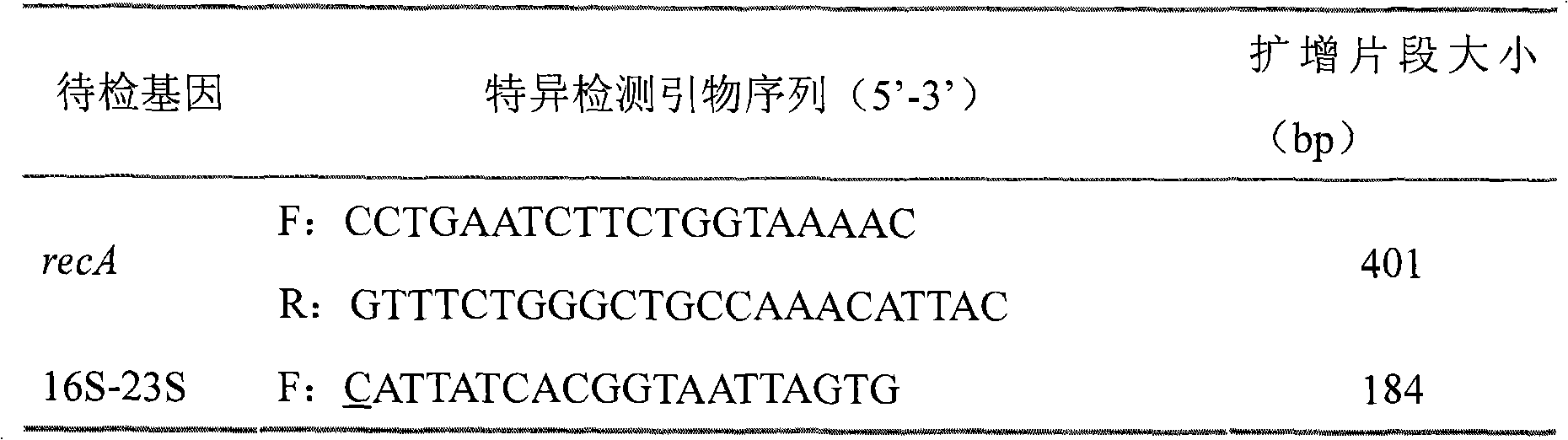
[0076] Ab complete genome sequence information was obtained from GenBank. Three specific detection segments were determined, and the genes anchored by each segment involved the recA gene, the 16S-23S ITS region, and the OXA-51-like gene, and were detected by mPCR to improve the specificity of Ab identification.
[0077] 3. Select specific PCR primers for the three detection segments, a...
Embodiment 2
[0091] Example 2 Application of mPCR detection in multiple detection objects
[0092] This example mainly illustrates how the mPCR system uses a variety of specific detection primers to identify unknown samples. That is, the detection of Streptococcus pneumoniae (Sp), Neisseria meningitidis (Nm) and Listeria monocytogenes (Lm) in clinical cerebrospinal fluid specimens is the main cause of meningitis pathogenic bacteria. The main difference between this example and Example 1 is that for complex samples (containing multiple nucleic acid sequences to be detected, or multiple species), 3 detection target fragments are set for each nucleic acid sequence of the species to be detected, involving 3 For the UT+specific detection primer complex, the UT sequence is the same; for different species to be tested, there are different UT primer pairs to show the difference, but the Tm values of each UT are similar, and the mPCR reaction conditions are uniform. In this way, the vertical di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com