Polypeptide pair for specifically recognizing muscle myostatin gene as well as encoding gene and application of gene
A muscle growth inhibition, one-to-one technology, applied in the fields of peptides, depsipeptides, DNA preparation, etc., can solve the problems of inability to guarantee gene knockout, time-consuming, cumbersome ZFN preparation process, etc., and achieve the effect of great application value.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1, the construction of TALE target recognition module
[0045] 1. Description of the four modules and the mechanism of combining modules
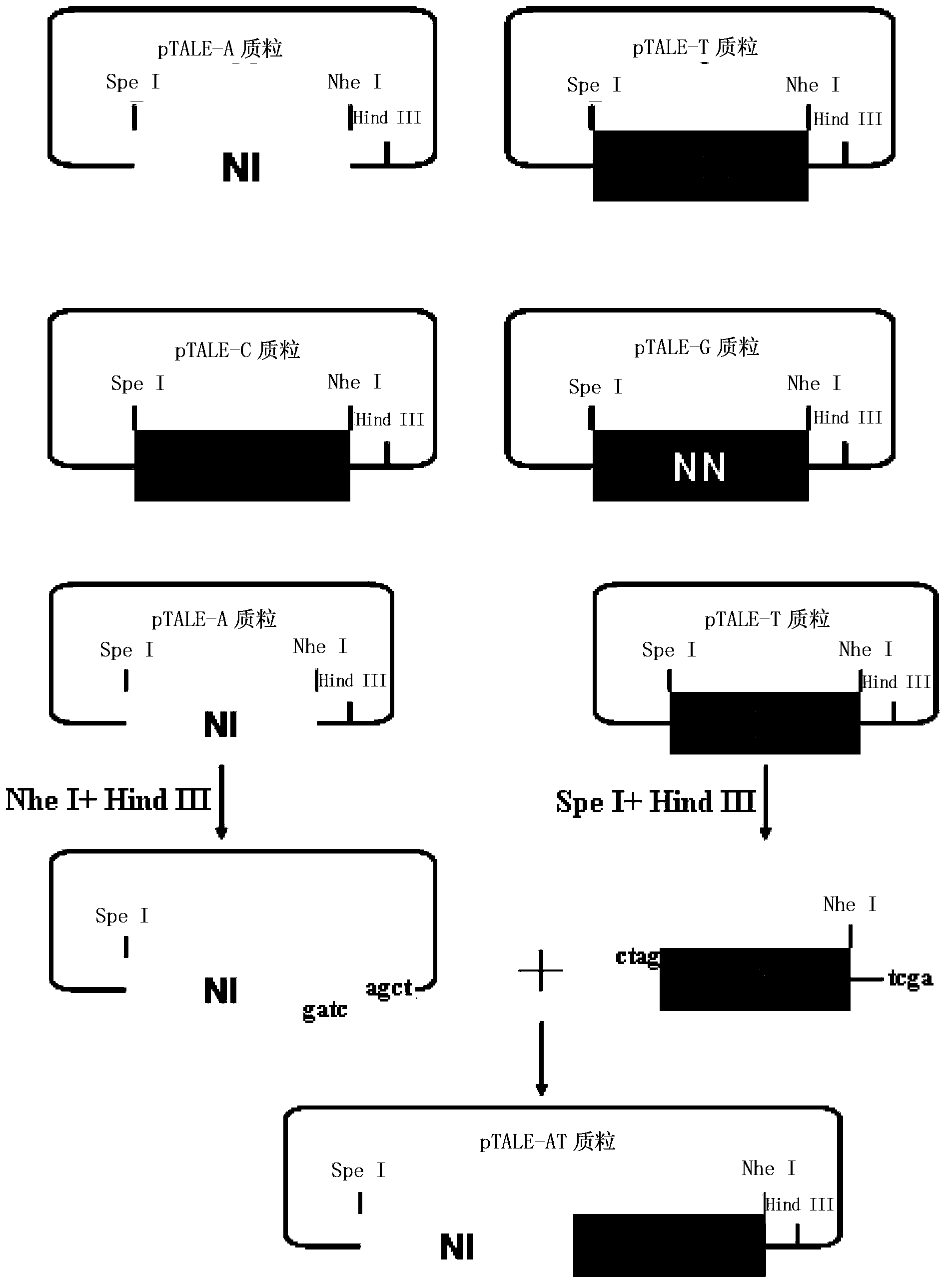
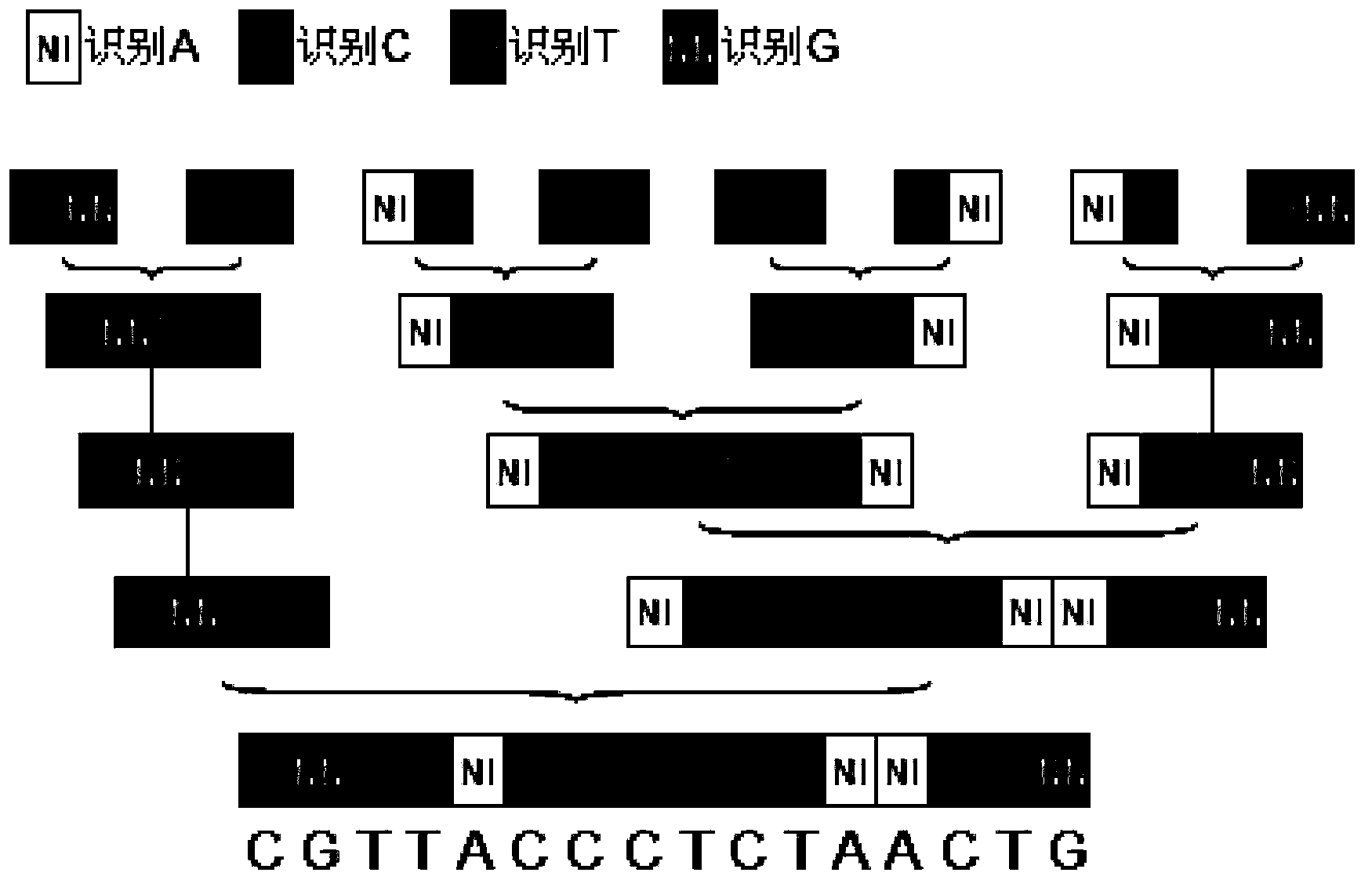
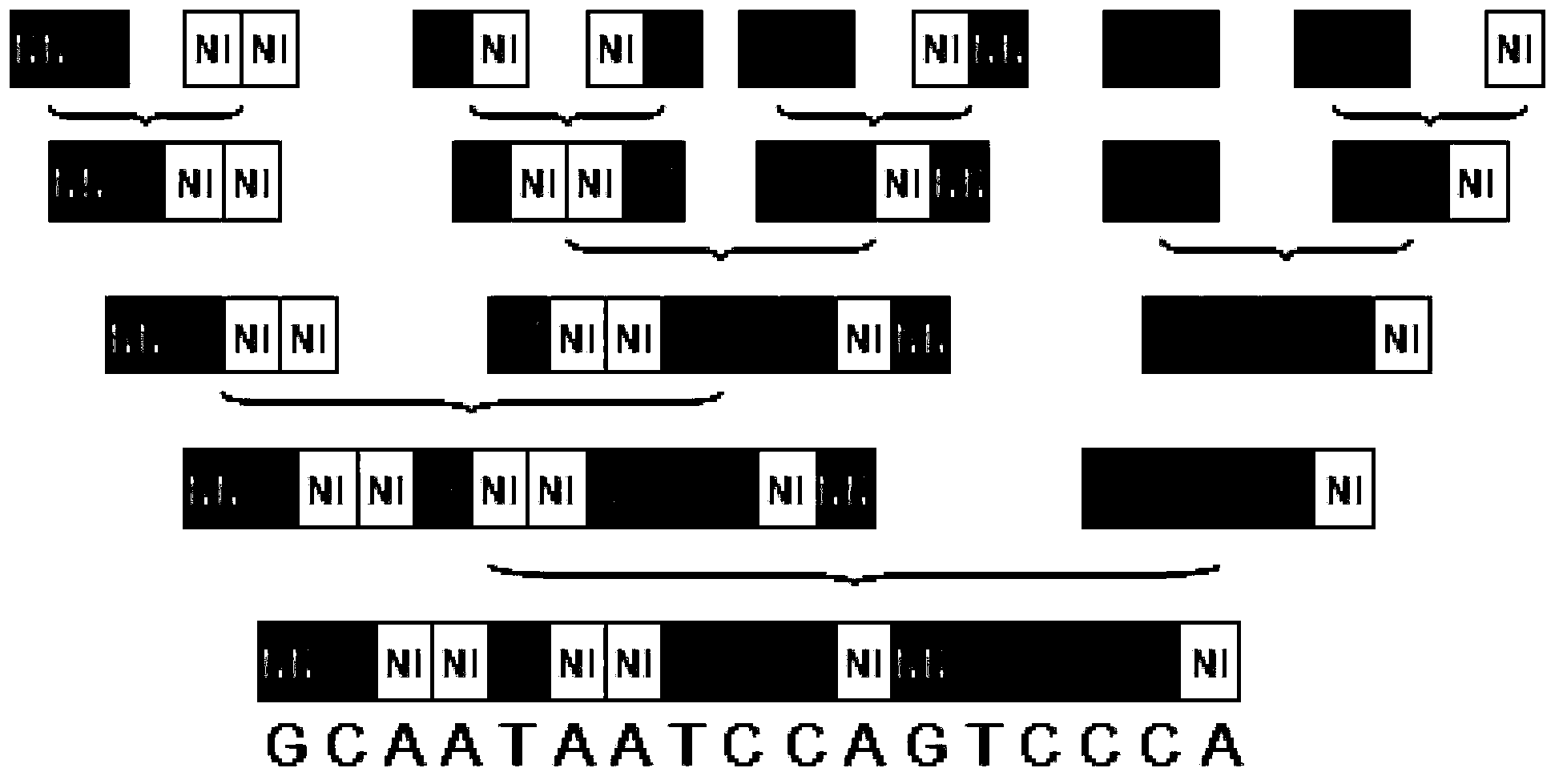
[0046] The nucleic acid recognition unit of TAL is a double-linked amino acid separated by 32 constant amino acid sequences. Double-linked amino acids have a constant correspondence with A, G, C, and T, that is, NI recognizes A, NG recognizes T, HD recognizes C, and NN recognizes G. The pTALE-A plasmid, pTALE-G plasmid, pTALE-C plasmid, and pTALE-T plasmid are single-module vectors, which are plasmids encoding DNA for the nucleic acid recognition units of the above four TALs respectively, and at the 5' end of the encoding DNA. Spe I restriction recognition sequence, 3' end has continuous Nhe I restriction recognition sequence and Hind III recognition sequence. Through the Spe I, Nhe I, and Hind III restriction sites on the single-module vector, the TAL unit corresponding to the target sequence can be cloned in series, an...
Embodiment 2
[0065] Embodiment 2, the construction of reporter plasmid
[0066] 1. Synthesize the double-stranded DNA molecule (target fragment) shown in sequence 6 of the sequence listing.
[0067] 2. Using the double-stranded DNA molecule synthesized in step 1 as a template, perform PCR amplification with a primer pair composed of mstnssaup and mstnssadn to obtain a PCR amplification product.
[0068] mstnssaup:5'-AGT AGATCT GTGATTCAGGATCTATTGCTAC-3';
[0069] mstnssadn:5'-AGT CTCGAG GCAGTAATTGGCCTTATATCT-3'.
[0070] 3. The PCR amplified product of step 2 was double-digested with restriction endonucleases BglII and XhoI, and the digested product was recovered.
[0071] 4. Digest the pSSA vector with restriction endonucleases BglII and XhoI to recover a vector backbone of about 6500 bp.
[0072] 5. Ligate the digested product of step 3 with the vector backbone of step 4 to obtain the recombinant plasmid pSSA-MSTN (reporter plasmid). According to the sequencing results, the struct...
Embodiment 3
[0073] Example 3, verifying the TALEN activity of the plasmid constructed in Example 1 by the luciferase reporter gene method
[0074] The following two groups of experimental treatments were carried out respectively (each experimental treatment was repeated three times, and each repeated experiment was set with three repeated treatments, and the average value of the three repeated treatments was taken for the results; the transfection reagents were all DNA Fect Transfection Reagent DNA transfection reagents, CWBIO , Cat No.CW0860, the amount of transfection reagent added in each treatment is 6ul, and the operation is performed according to the instructions):
[0075] Group 1: 0.4ug Renilla luciferase plasmid (reference control plasmid) and 2.0ug recombinant plasmid pSSA-MSTN were co-transfected 1×10 6 293T cells;
[0076] Group 2: Co-transfect 1× with 0.4ug Renilla luciferase plasmid (reference control plasmid), 2.0ug recombinant plasmid pSSA-MSTN, 4ug recombinant plasmid pc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com