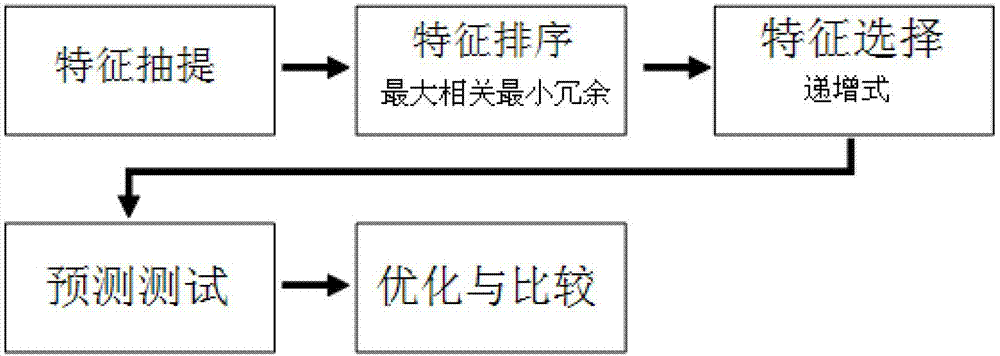
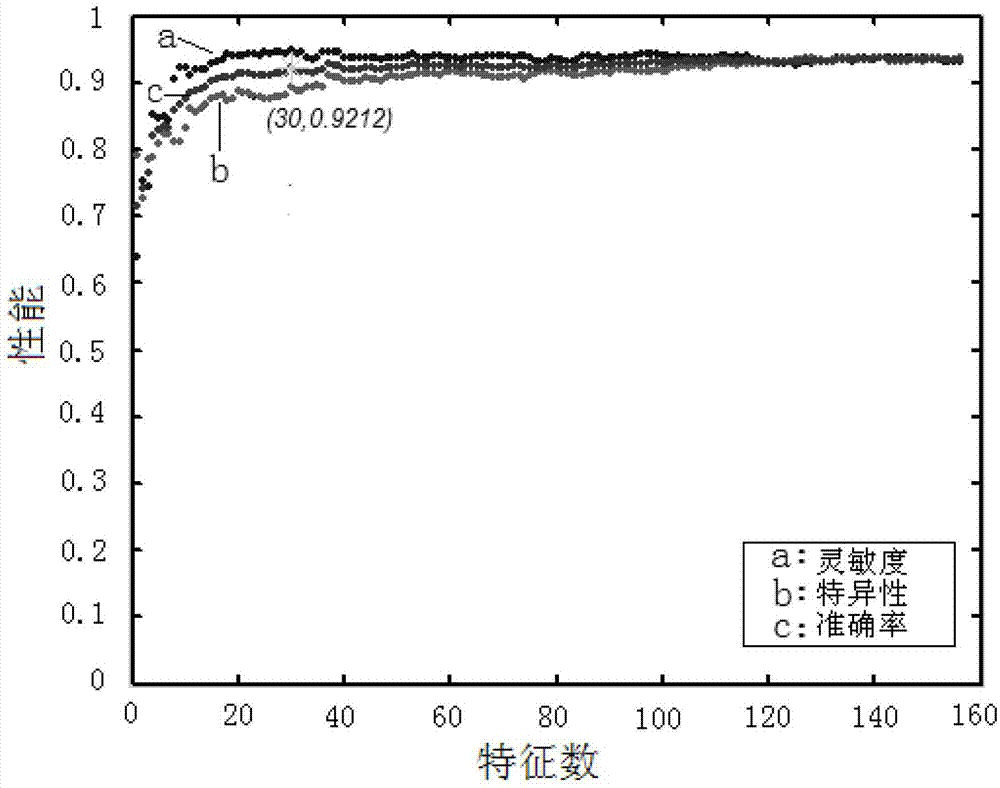
Method for predicting potential sensitization in protein
A prediction method, protein technology, applied in the field of computational biology prediction of protein characteristics, can solve the problems of high false positive rate, failure to meet actual needs, and inability to know allergen characteristic information, etc., to achieve the improvement of sensitivity and specificity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Embodiment 1, Dataset Preparation for Prediction Methods
[0025] Allergen protein dataset preparation: data source databases include Swiss-Prot Allergen Index (an authoritative protein database); IUIS Allergen Nomenclature (International Union of Immunological Societies-Allergen Nomenclature); SDAP (allergen structure database) and ADFS (using Allergen database for food safety), a total of 1176 allergen protein sequences were obtained after removing redundancy, and these sequences were used as the positive set for training the model;
[0026] Preparation of non-allergen protein data set: In order to construct a feasible negative set, the following steps are taken: 1. Download all protein sequences of Swiss-Prot (version: 2010_11), a total of 522,019; 2. Remove all known allergens Sequences with a similarity >= 30%; 3. Remove all sequences with a length less than 50 amino acids; 4. Randomly select protein sequences with the same number as the positive set from the res...
Embodiment 2
[0027] Embodiment 2, Input feature vector construction of SVM
[0028] Physicochemical characteristics: The physical and chemical characteristics of the protein collected in this example include eight aspects, 1. Amino acid composition; 2. Molecular weight; 3. Secondary structure tendency; 4. Hydrophobicity; 5. Polarization; 6. Solubility; 7. Normalized van der Waals volume; 8. Polarity; 9. Sequence length. The formula for amino acid composition is Fraction of a min o acid i = total number of a min o acids ( i ) total number of a min o acids in protein ; In addition to amino acid composition, molecular weight and sequence length, the other six attributes are related to a single amino acid and can be divided into 2 to 3 catego...
Embodiment 3
[0034] Embodiment 3, Feature selection for building models
[0035] The mRMR (Maximum Relevance Minimal Redundancy) method is used to sort the features before modeling, and the parameters of the mRMR program are set to λ=1, m=MID. The sorted feature list is used in the subsequent incremental feature selection process;
[0036]Use the IFS (Incremental Feature Selection) process to model, evaluate performance and obtain the optimal feature composition. The specific principle is to first select the top-ranked feature for modeling, and calculate its performance parameters for 10-fold cross-validation , and then select the top 2 features for modeling, and calculate the performance parameters of its 10-fold cross-validation, and so on, each time adding a top feature, until all the features are added, a total of the same For performance parameter pairs with the same number of features, draw a performance curve. The performance curve obtained by this method gradually rises from lo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com