Gene marker for detecting cancer tissue and para-carcinoma tissue of primary liver cancer
A gene chip, specific technology, applied in the field of medicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0034] The preparation of the gene chip of the present invention can adopt the conventional manufacturing method of biochip. For example, if the solid phase carrier is a modified glass slide or a silicon wafer, and the 5' end of the probe contains amino-modified poly dT strings, the oligonucleotide probe can be formulated into a solution, and then the spotting instrument will spot it on The gene chip of the present invention can be obtained by modifying glass slides or silicon slices, arranging them into a predetermined sequence or array, and then fixing them by placing them overnight. If the oligonucleotide probe does not contain amino modification, its preparation method can also refer to: "Gene Diagnosis Technology-Non-radioactive Operation Manual" edited by Wang Shenwu; J.L.erisi, V.R.Iyer, P.O.BROWN.Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997; 278: 680 and Ma Liren, edited by Jiang Zhonghua. Biochip. Beijing: Chemical I...
Embodiment 1
[0051] Embodiment 1, the preparation of RNA sample
[0052] Tissue sample:
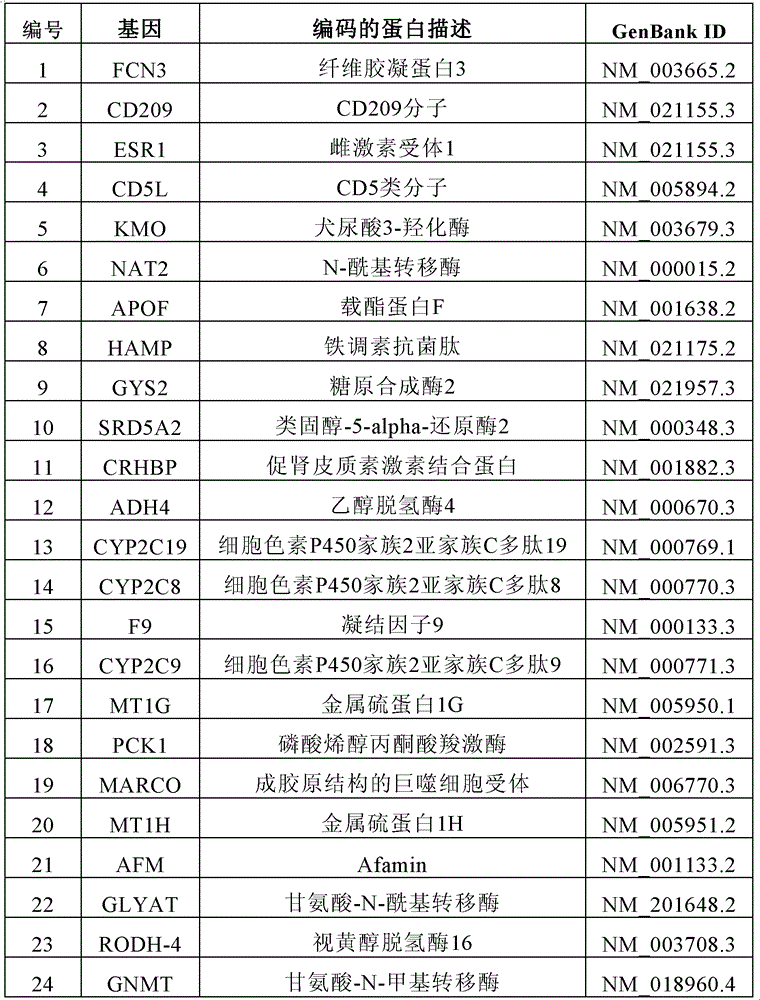
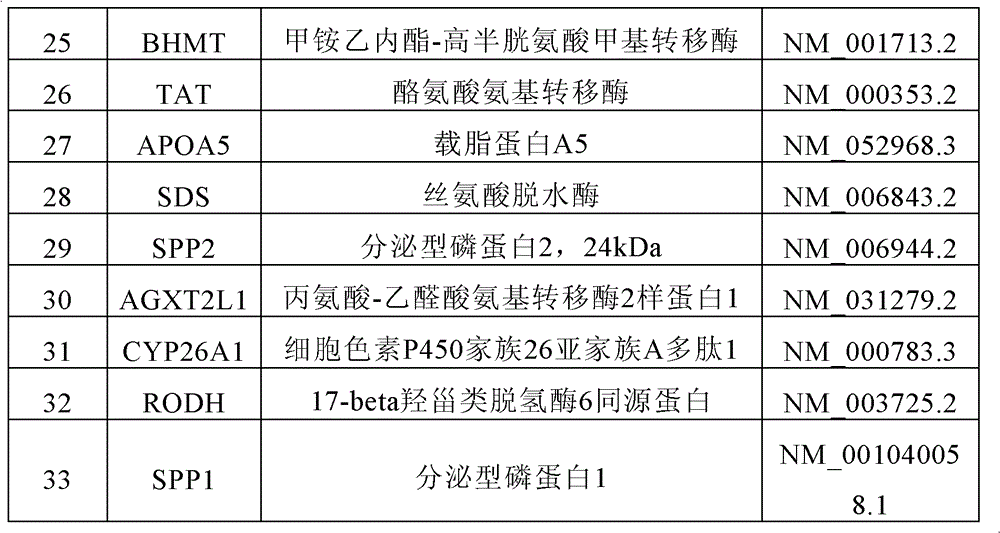
[0053] Forty-nine pairs of cancer and paracancerous tissues were obtained from surgical resection specimens of patients with primary liver cancer, and these specimens were obtained from Qidong Liver Cancer Institute and the First Affiliated Hospital of Zhejiang University. 10 cases of normal liver from accidental death. All the above-mentioned specimens were obtained with the approval of the ethics committee of the WHO partner organization authorized by the Shanghai Municipal Government. The clinical data of tissue samples include: gender, age, tumor size, pathological grade (Edmonson), liver cirrhosis, metastasis, recurrence, HBV and HCV infection, etc. All patients did not receive chemotherapy before operation, and the postoperative follow-up was up to 60 months.
[0054] Gene chip:
[0055] Whole-genome expression profile chip, using the crystal core of Capital Bio Co., Ltd. (http: / / www.capital...
Embodiment 2
[0061] Example 2, screening genes with significant differences
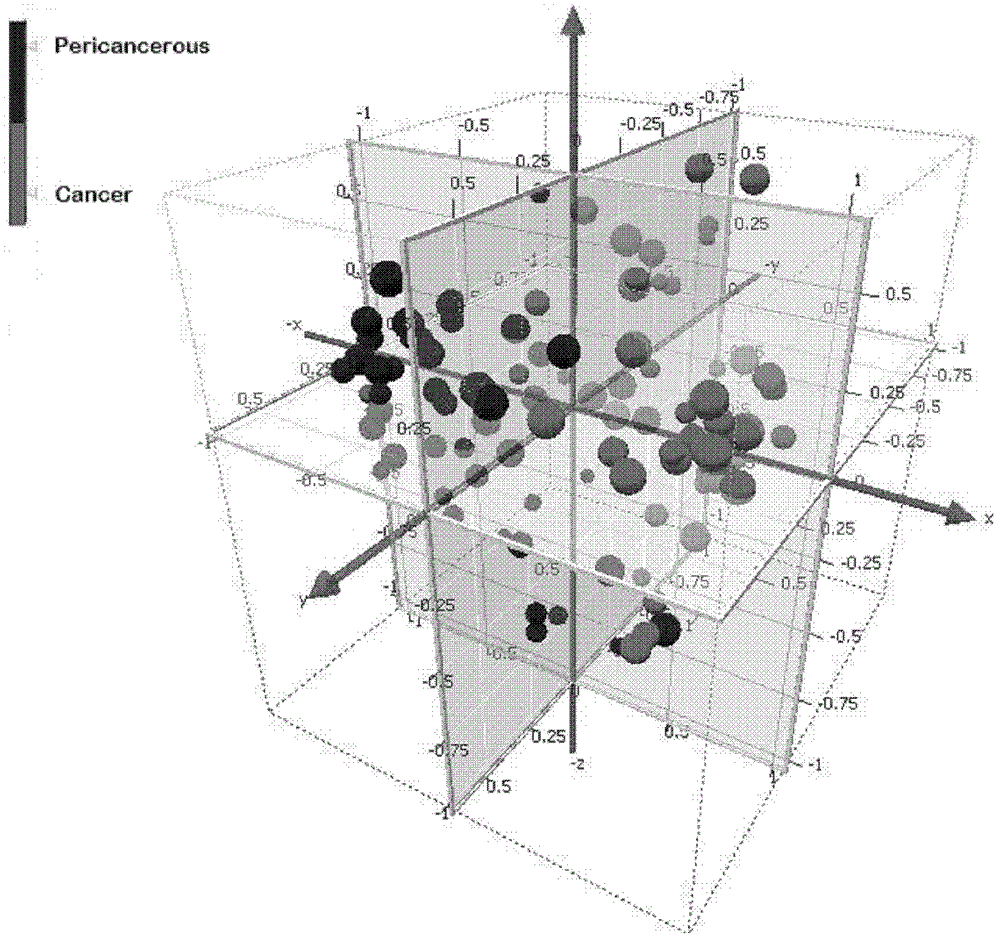
[0062] 49 cases of cancer samples and normal samples (10 cases mixed), 49 cases of paracancerous samples and normal samples (10 cases mixed) were labeled with two fluorescent dyes, cy3 and cy5, respectively, and competitively hybridized with the probes on the genome-wide expression profile chip . Chip core after hybridization LuxScan TM The 10K microarray chip scanner scans to obtain the result image, and then quantifies the hybridization result through the LuxScan 3.0 general microarray image analysis software that comes with it. Thus, the expression profile data of 49 pairs of cancer samples to normal samples (cancer / normal, Cancer / Normal, C / N) and para-cancer samples to normal samples (pre-cancer / normal, Pericancerous / Normal, P / N) were obtained.
[0063] The above 49 cancer / normal microarray results and 49 paracancerous / normal microarray results were first normalized by LOWESS (local weighted regression a...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap