Efficient assembling method of transcription activator-like effectors (TALE) repeating region for editing silkworm genome and framework carrier thereof
An assembly method and genome technology, applied in the field of molecular biology, can solve problems such as increased economic costs, difficult synthesis, unfavorable gene editing, etc., and achieve the effects of cost reduction, rapid and effective assembly, and convenient editing and transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] In order to elaborate the principle of the present invention in detail, it will be described below in conjunction with actual results and accompanying drawings. For the experimental methods not specified in the examples, the conventional conditions are generally followed, such as the conditions described in the Molecular Cloning Experiment Guide (Third Edition, J. Sambrook et al.), or the conditions suggested by the manufacturer.
[0034] The silkworm cell line used in the examples is the silkworm embryo cell line BmE, cultivated and preserved by the State Key Laboratory of Silkworm Genome Biology, Southwest University, China.
[0035] 1. Principle and method of TALE assembly suitable for editing silkworm genome
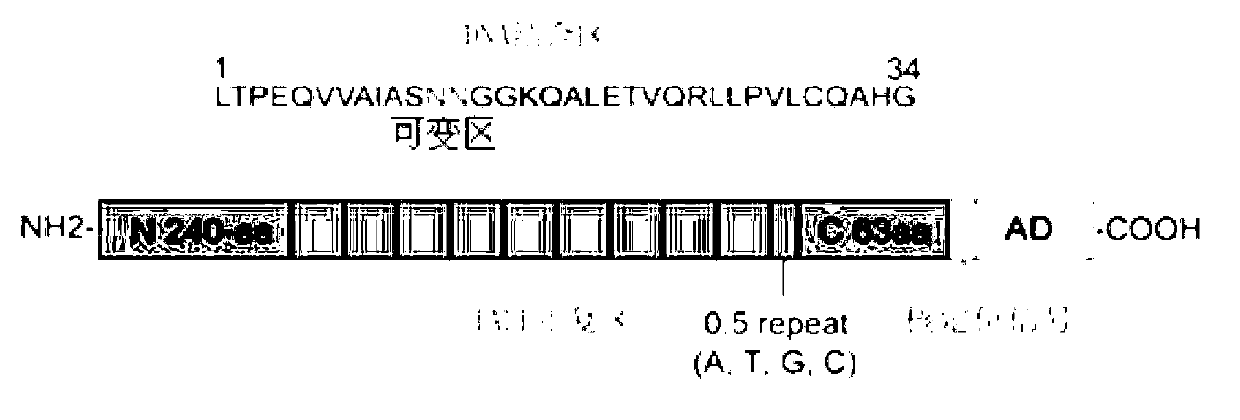
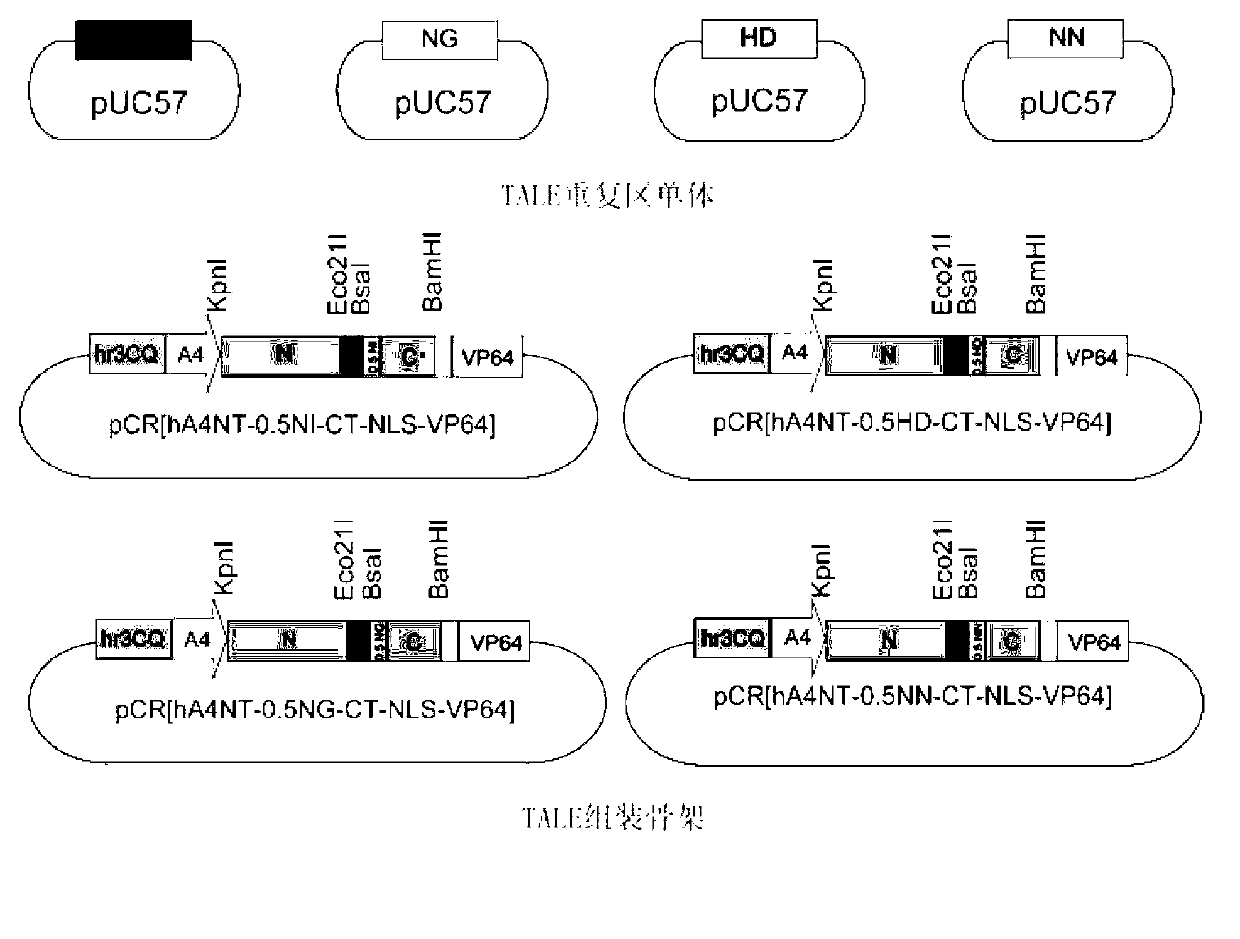
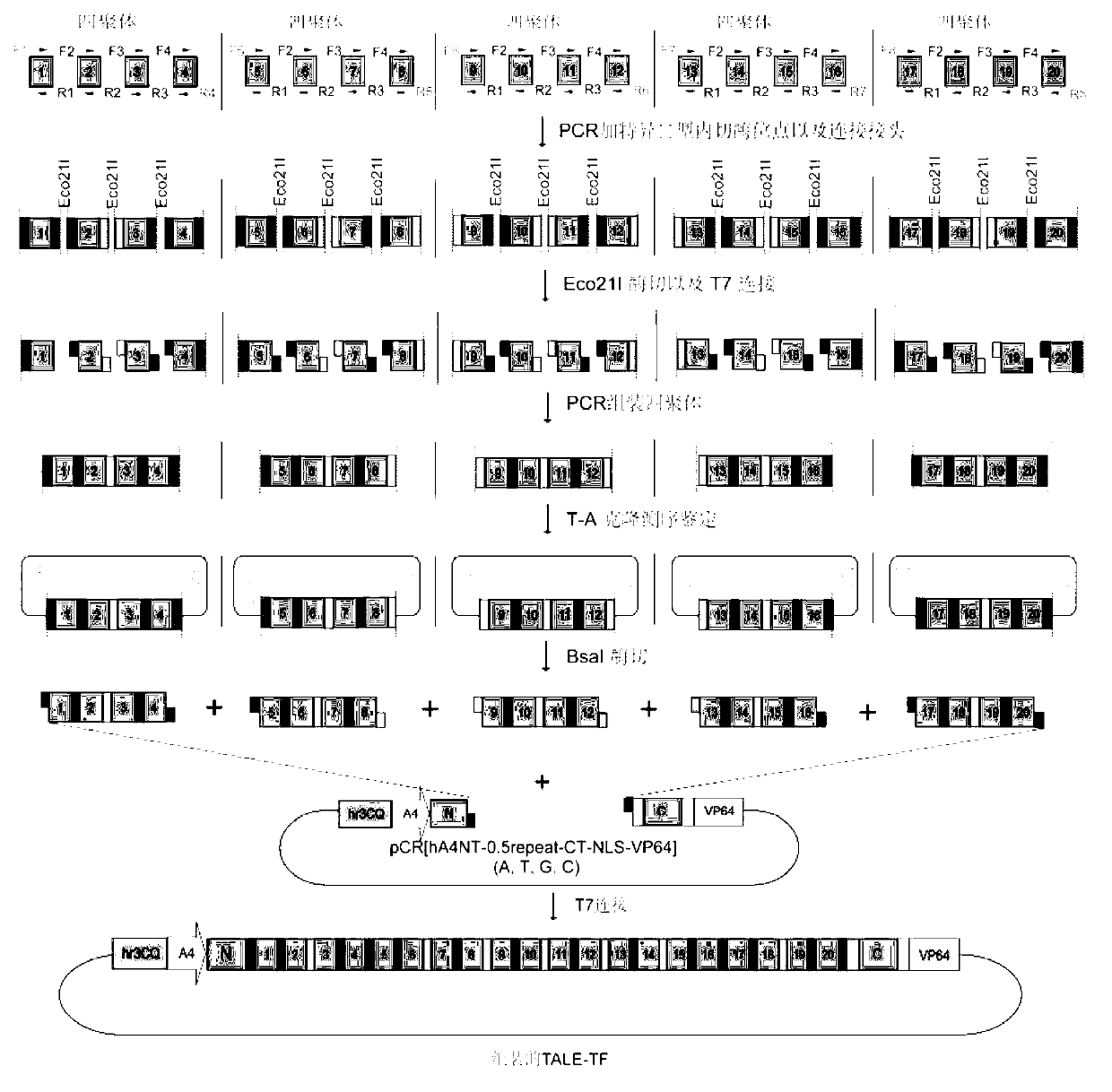
[0036] TALE consists of N-terminal residues, several repeat regions in series, 0.5 repeat regions, C-terminal residues, nuclear localization signals and transcriptional activators, such as figure 1 shown. TALE can specifically recognize and bind target DNA t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com