Method for identifying drought resistance of highland barley by utilizing semi-quantitative PCR method of HVA1 gene
A drought resistance, semi-quantitative technology, applied in the field of identification of drought resistance of highland barley, can solve the problems of plant drought resistance without physiological and biochemical indicators, cumbersome methods, and difficult identification of plant drought resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0017] The present invention will be described in detail below in conjunction with specific embodiments.
[0018] Table 1 Primer name and sequence
[0019]
[0020] Primer P1 was designed according to the sequence of barley HVA1 gene (X78205) published by GenBank, and a pair of primers P2 were designed for barley actin (β-Actin) gene (ID: AY145541) using Primer5.0. synthesis.
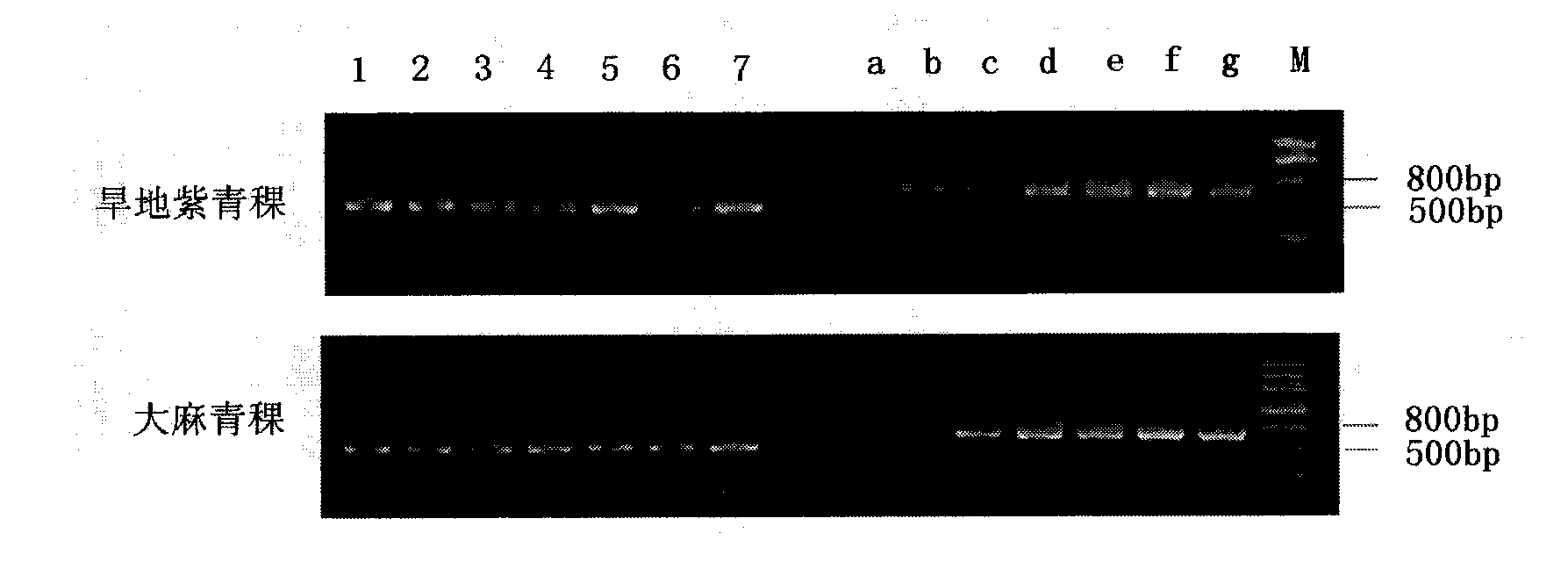
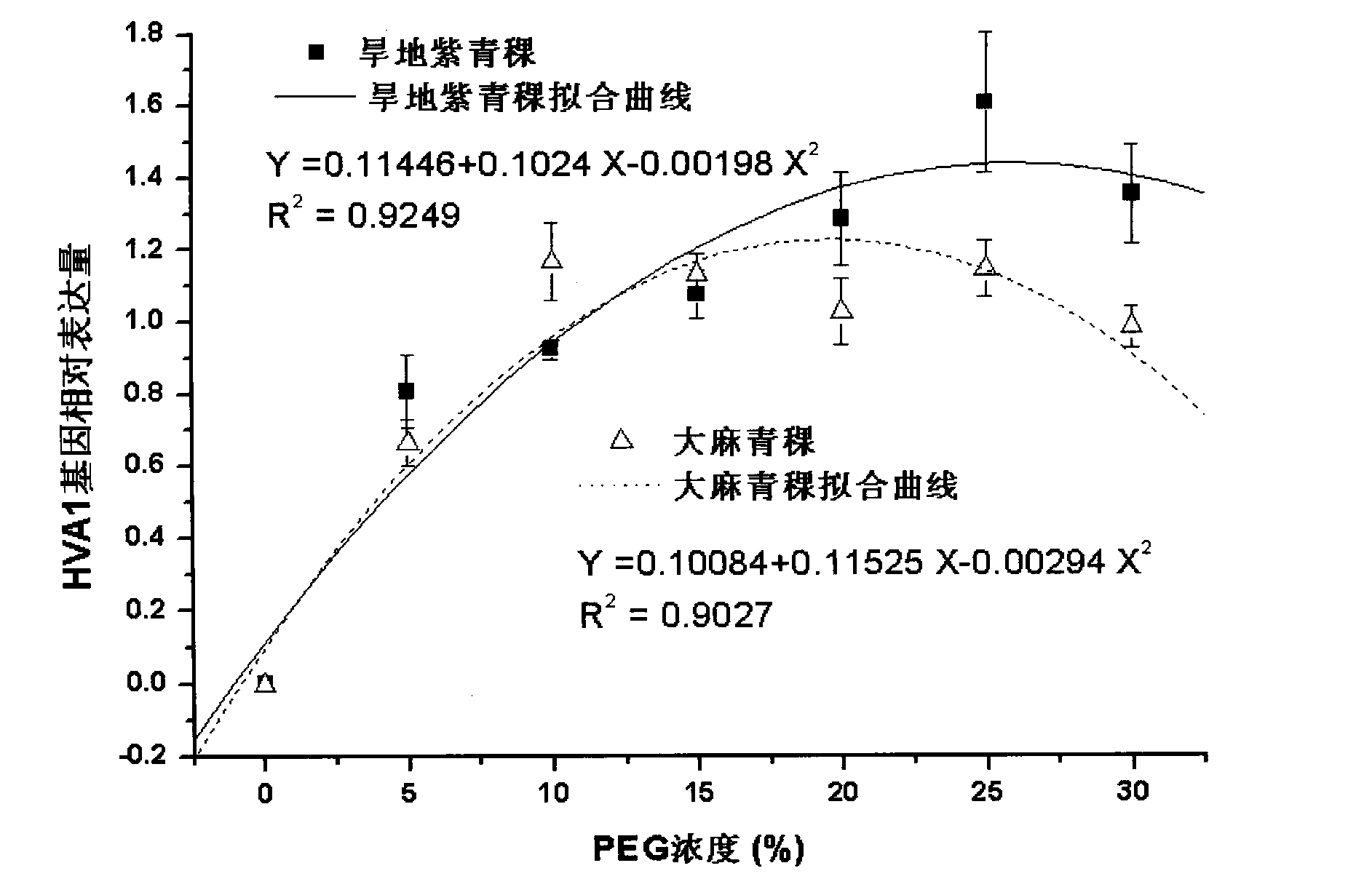
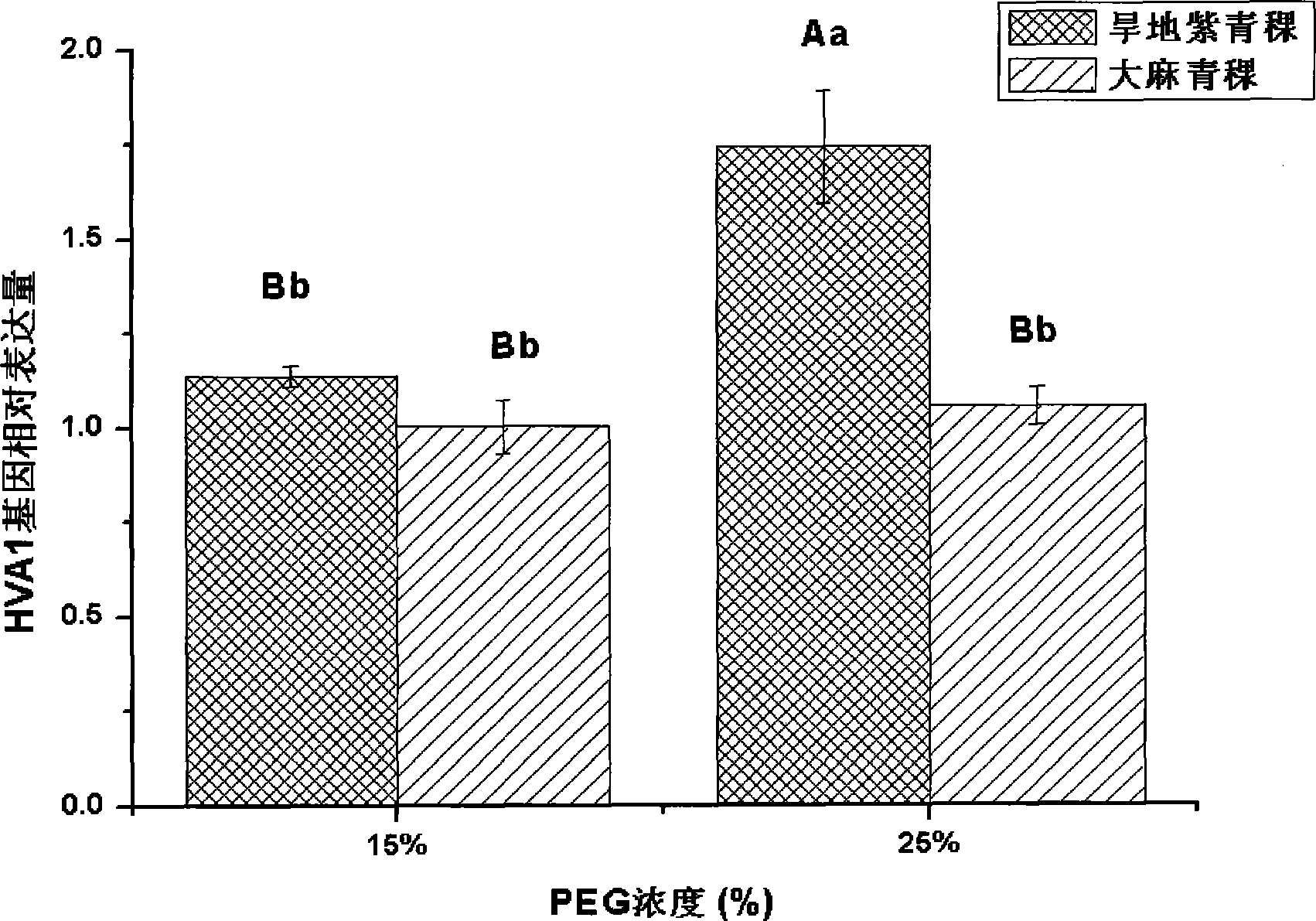
[0021] Using drought-resistant purple highland barley and drought-sensitive hemp highland barley as materials, select highland barley seeds with consistent plumpness and no damage from diseases and insect pests (two varieties with strong drought resistance and weak drought resistance), soak in clean water for 1 hour, and use 0.5% high Potassium manganate was sterilized for 30 minutes, washed with water, and the seeds were immersed in distilled water and placed in a refrigerator at 4°C for 24 hours. Take 20 seeds each and sow them in a petri dish with a diameter of 10 cm, covered with 3 layers of di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com