High efficient expression method for actinomyces-based nitrile hydratase gene in escherichia coli
A technology of Escherichia coli and nitrile hydratase, applied in the field of microbial genetic engineering, can solve the problems of slow progress of nitrile hydratase gene research and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
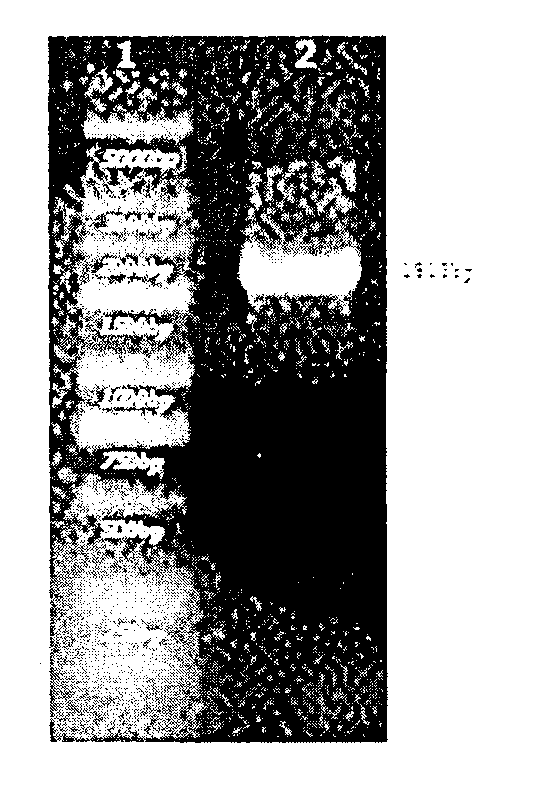
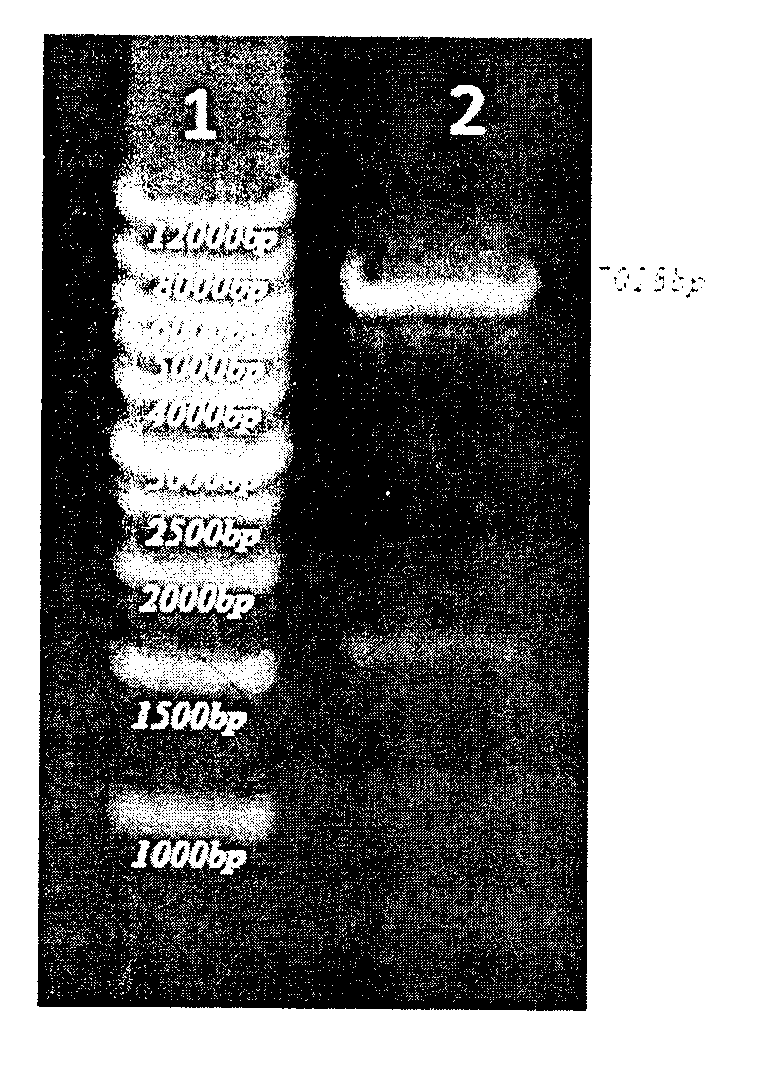
[0030] 1) Obtain the full-length gene of nitrile hydratase by PCR
[0031] Primers (BAE up, BAE down) were designed to amplify the full-length nitrile hydratase gene sequence by referring to the nitrile hydratase gene sequence in Rhodococcus.
[0032] The upstream primer BAE up (containing the Nde I restriction site) is shown in SEQ ID NO.2, and the downstream primer BAE down (containing the HindIII restriction site) is shown in SEQ ID NO.3;
[0033] 2) Design a pair of primers with Escherichia coli SD sequence and spacer sequence
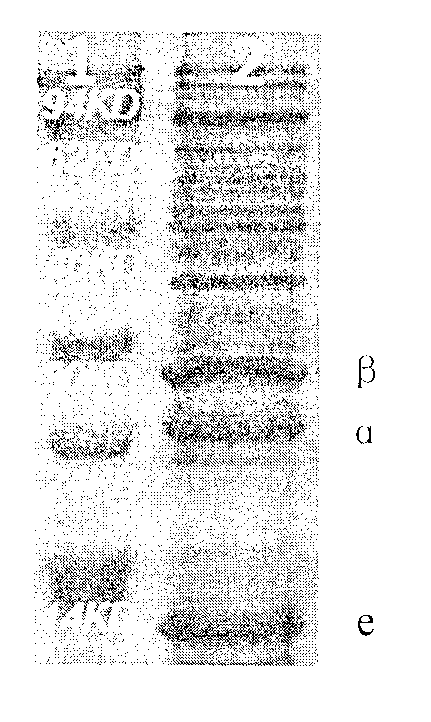
[0034] The complete sequence of the natural nitrile hydratase in Rhodococcus expressed in Escherichia coli BL21 has been reported internationally, but none of them are ideal. By replacing the upstream spacer sequence of the nitrile hydratase α gene and the regulatory protein e gene with SD sequence (as shown in SEQ ID 6) and a spacer sequence (as shown in SEQ ID 7), a large amount of soluble nitrile hydratase can be successfully expressed.
[003...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com