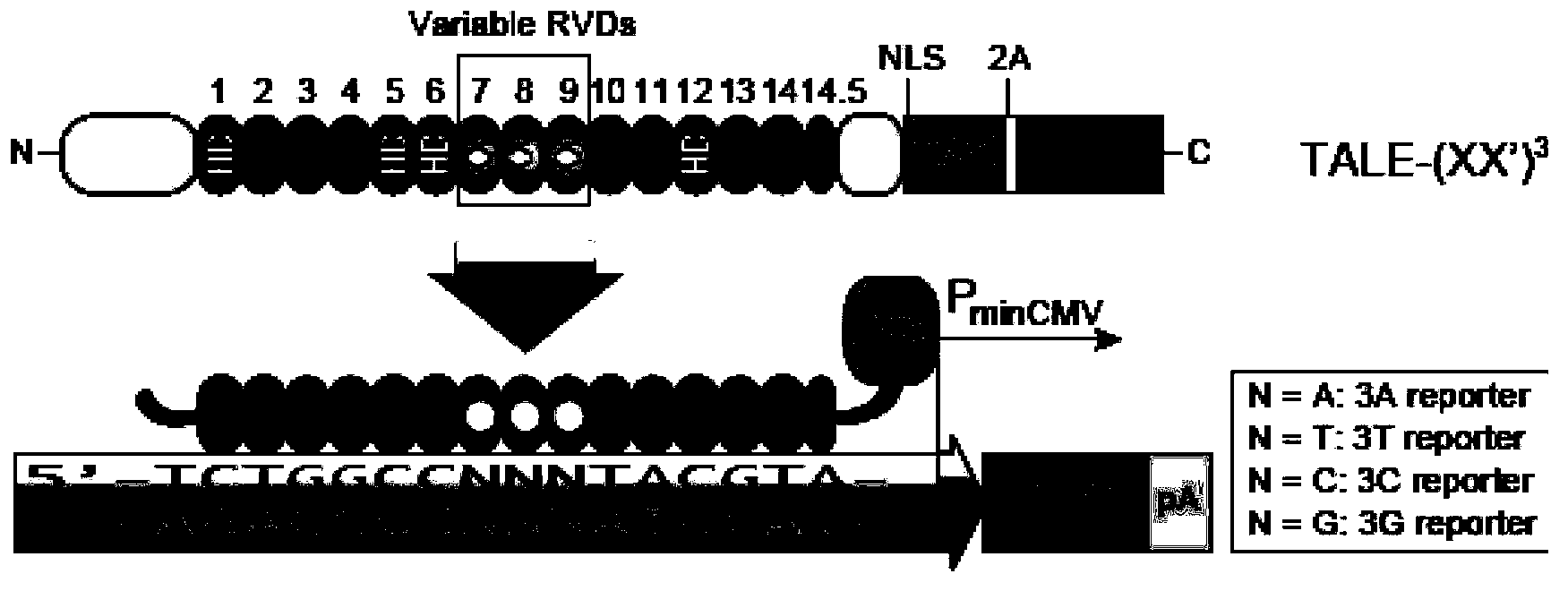
TALE (Transcription Activator Like Effectors) protein and application thereof
A protein and KG technology, applied in the field of genetic engineering, can solve the problems of reduced degree of freedom, low efficiency, RVD and DNA base specificity are not completely strict, etc., to improve specificity and efficiency, diversify selection, and reduce off-target effects. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 TALE transcriptional activator containing new RVD-KN
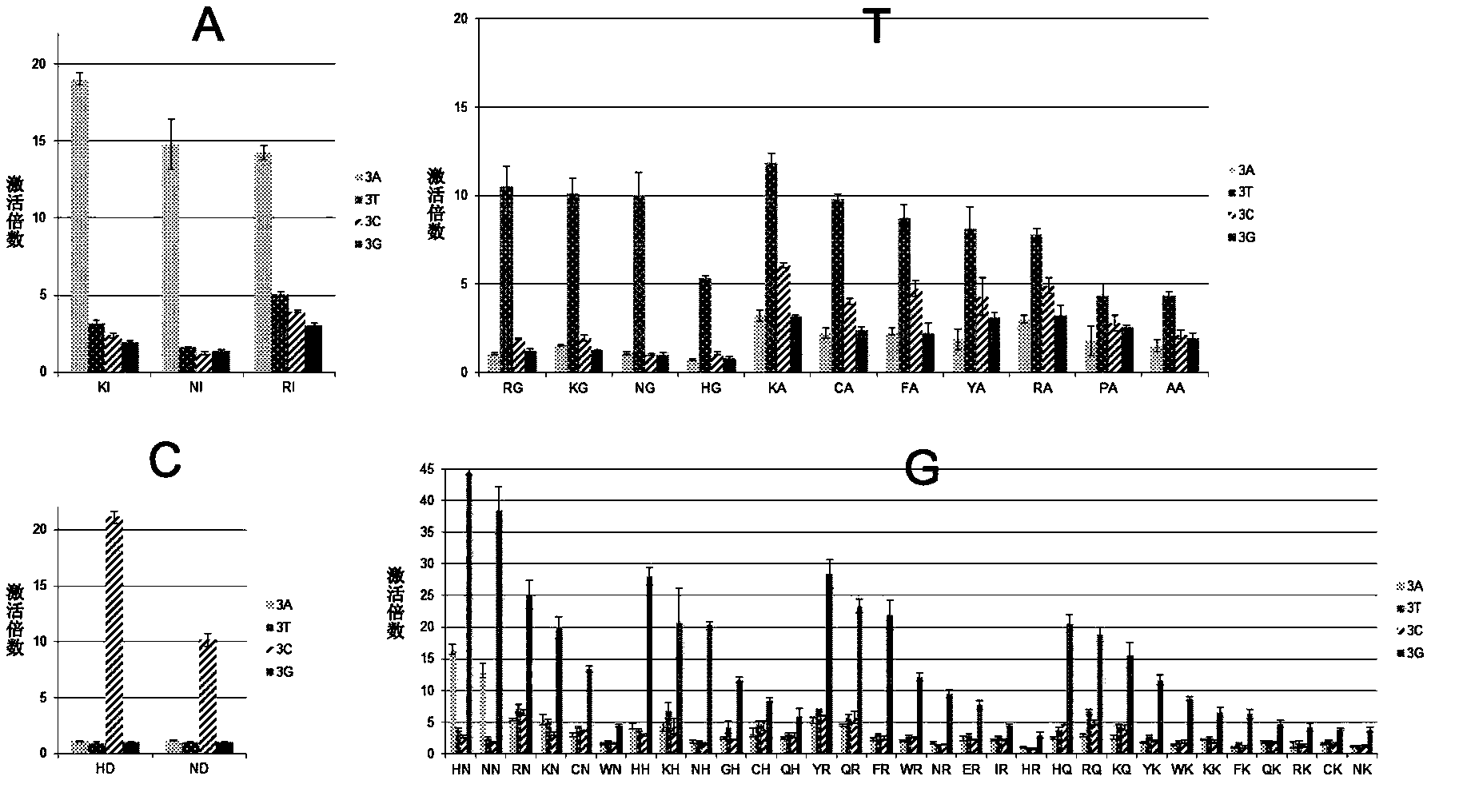
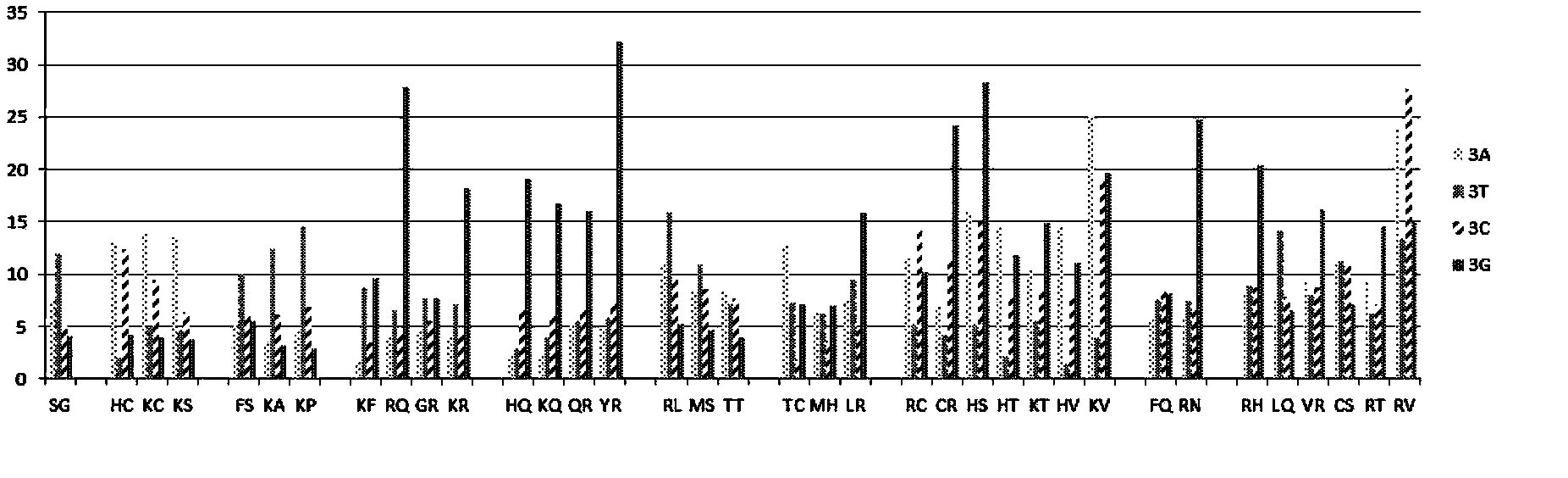
[0034] According to Table 1, KN is the RVD that specifically binds to the G base. For target site ACTC GGGGGGGGGGGG , Design a TALE-VP64 transcription activator with 15.5 repeats, and use RVD-----KN in the 5-15.5 repeats, and use the currently commonly used RVDs (NI-A, NI-A, NG-T, HD-C), the effect of activating the expression of the reporter gene downstream of the target site should be obtained. Since KN has specificity for G base binding, if the target site is replaced with ACTC AAAAAAAAAAAA , ACTC TTTTTTTTTTT T Or ACTC CCCCCCCCCCCC , Neither can activate the expression of downstream genes.
[0035] The constructed coding nucleotide sequence of the KN-containing TALE-VP64 transcription activator is shown in SEQ ID No. 1. The expression plasmid containing the KN TALE-VP64 gene (starting vector pLL3.7, purchased from Addgene, Inc.) and the reporter plasmid were co-transfected into the HEK293T cell line wi...
Embodiment 2
[0037] Example 2 TALE transcriptional activator containing novel RVD----RG
[0038] According to Table 1, RG is an RVD that specifically binds to the T base. For the target site CTGGCC TTT TACGTA, designed the TALE-VP64 transcription activator with 14.5 repeats, and used RVD in the seventh, eighth, and ninth repeats, and used the currently commonly used RVD (NI- A, NG-T, HD-C, NN-G), the effect of activating the downstream reporter gene expression of the target site should be obtained. Since RG has specific binding to T bases, if the target site is replaced with CTGGCC AAA TACGTA, CTGGCC CCC TACGTA or CTGGCC GGG TACGTA cannot activate the expression of downstream genes.
[0039] The coding nucleotide sequence of the constructed TALE-VP64 transcription activator containing RG is shown in SEQ ID No.2. The expression plasmid of the TALE-VP64 gene containing RG (starting vector pLL3.7) and the reporter plasmid were co-transfected into the HEK293T cell line with the transfection ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com