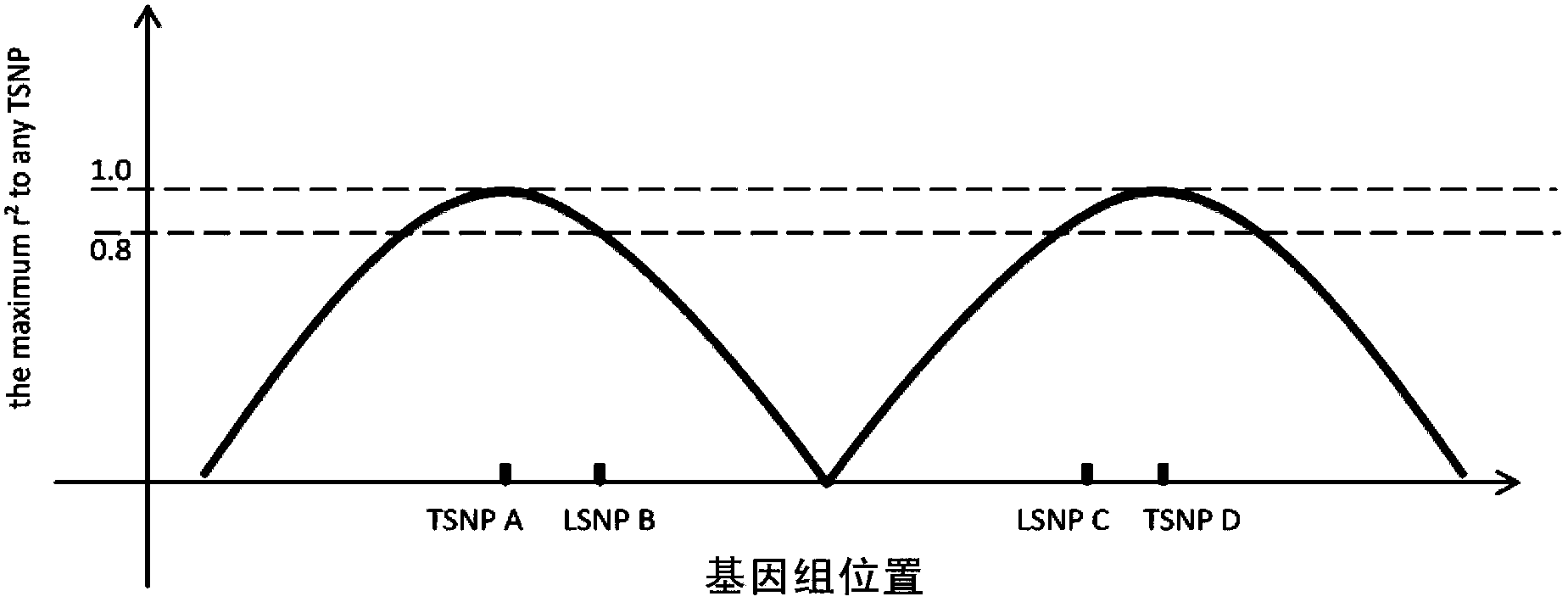
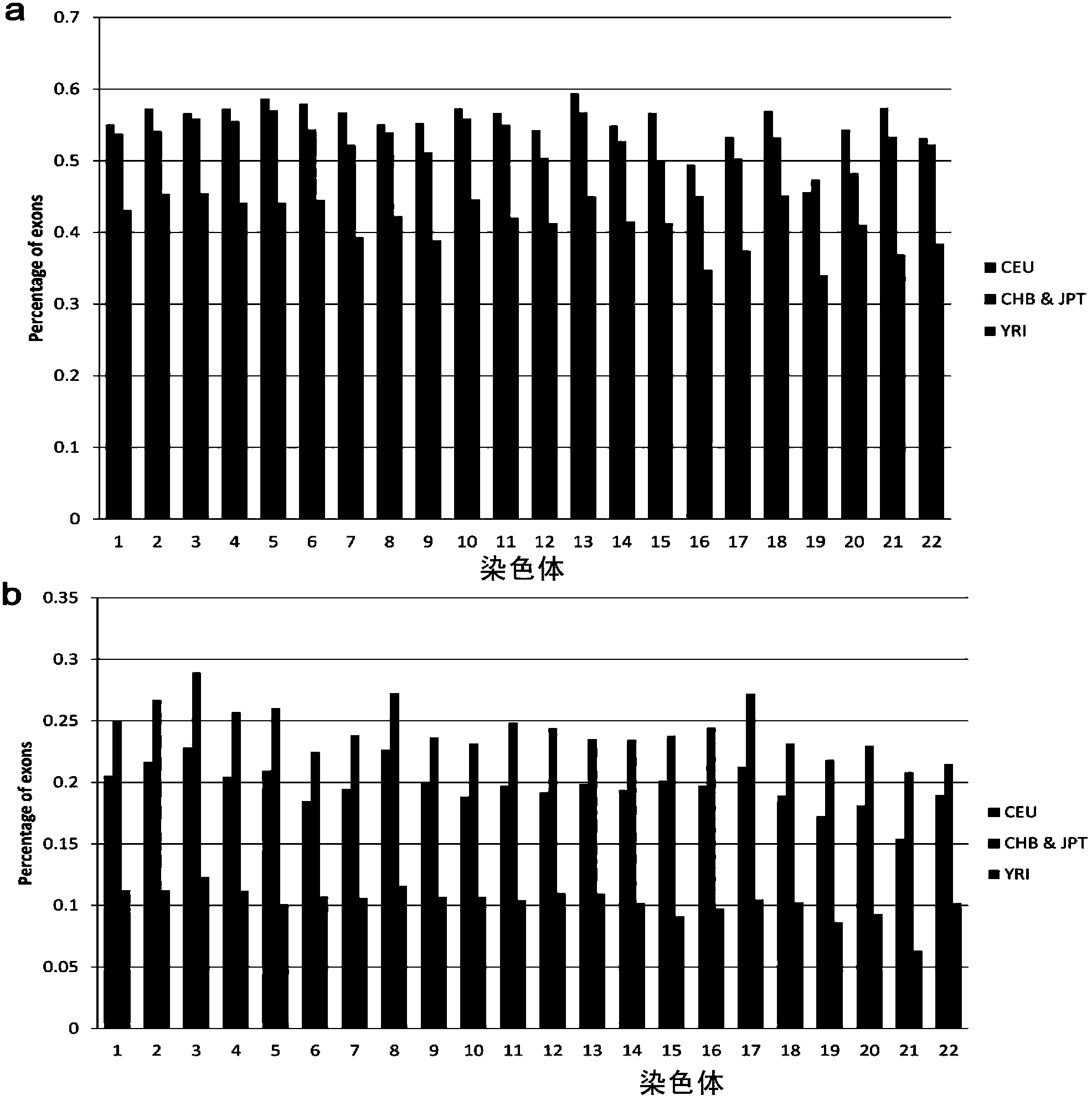
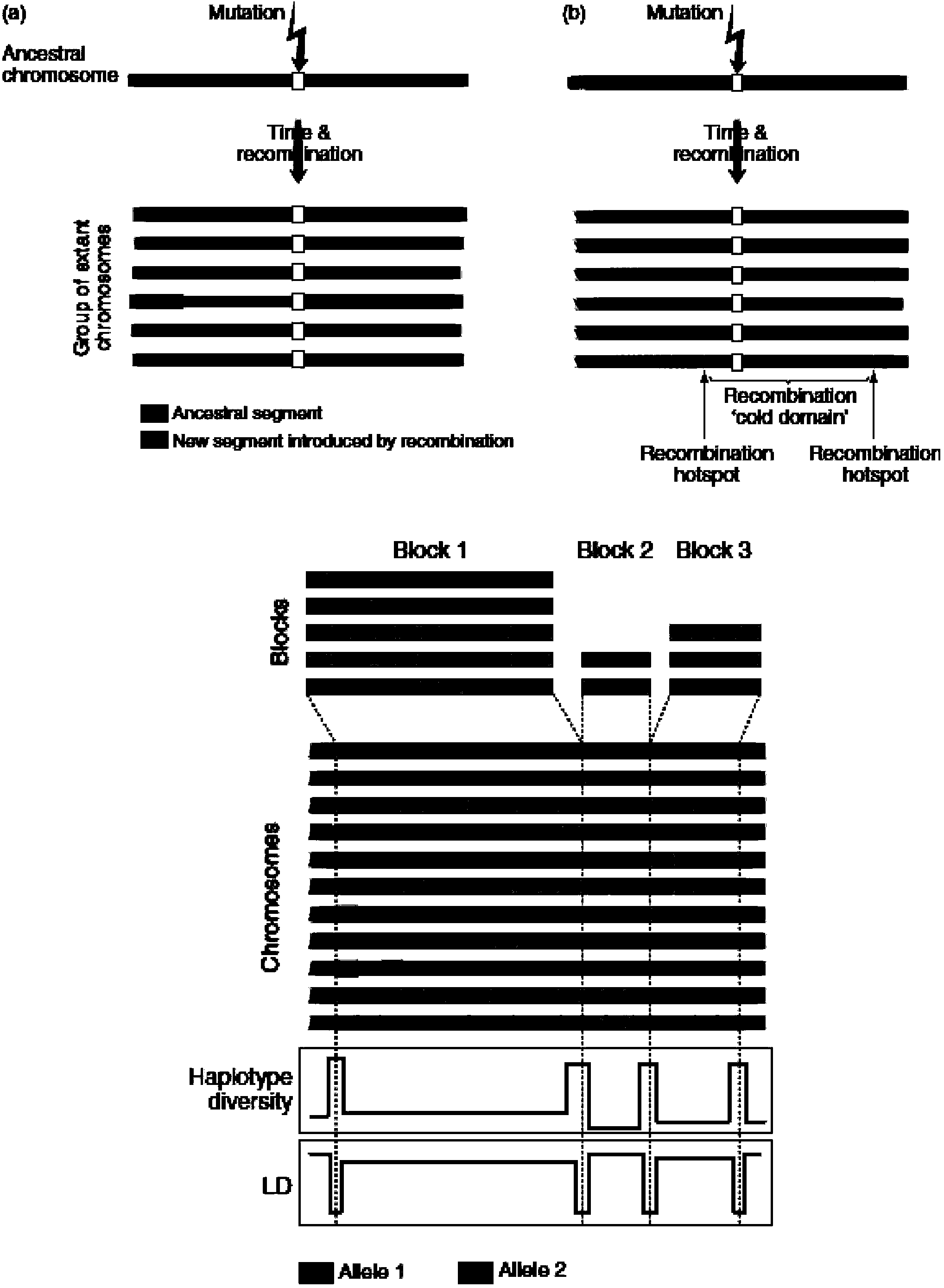
Method for detecting exome mononucleotide coverage rate
A single nucleotide and exon technology, applied in instruments, calculations, electronic digital data processing, etc., can solve the problems of questionable rationality and overestimation of coverage, and achieve the goal of reducing false positive rate and accuracy Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0072] Example 1: TXT file nearest_SNP_result.txt
[0073] run command
[0074] part 1='java -jar Haploview.jar-n-hapmapDownload-panel CEU-blockoutputGAB-png-infoTrack-dprime-release 27-check'
[0075] part2='-chromosome'+chromosome+'-startpos'+startpos+'-endpos'+endpos
[0076] part3='-out'+inputroot +' / '+word[0]+' / '+word[1]+' / '
[0077] output as Figure 5 . .LD file
[0078]
[0079]
[0080] .CHECK file
[0081]
[0082] Results and discussion
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com