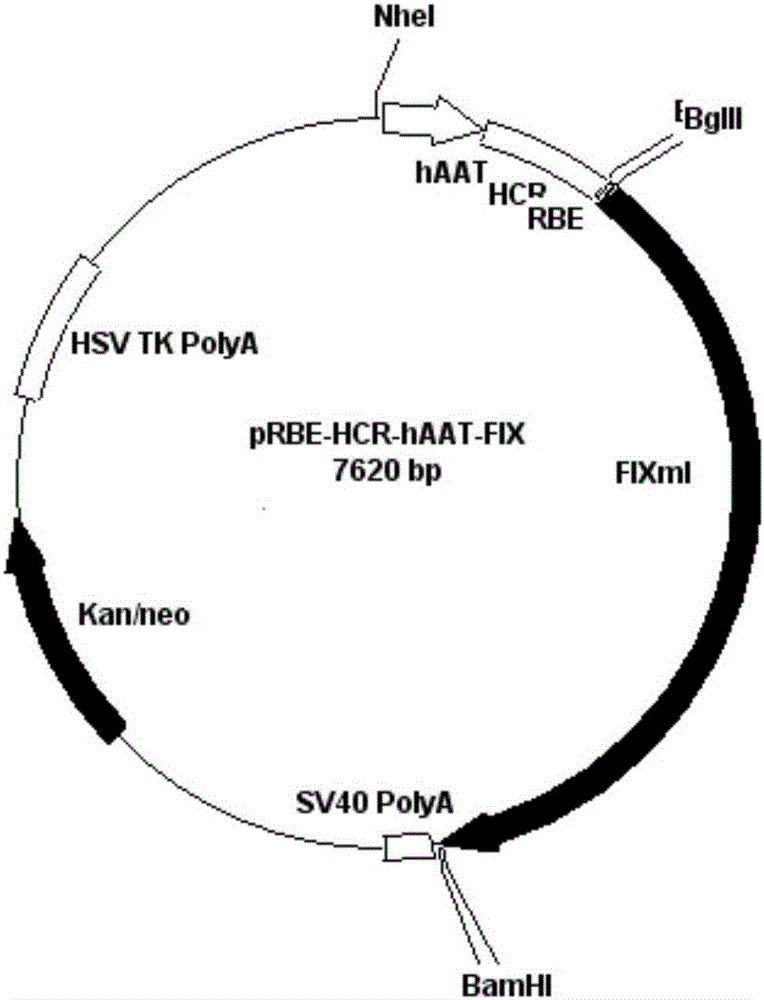
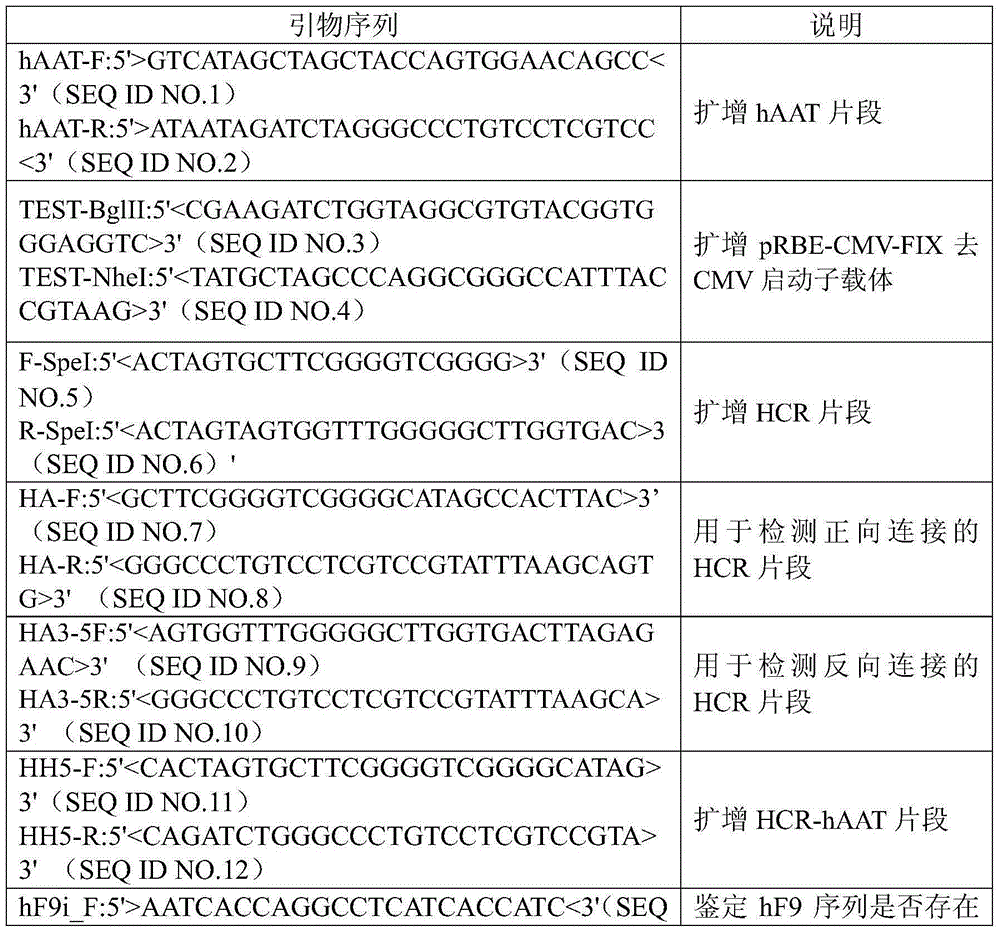
A kind of prbe-hcr-haat-hfixml plasmid and its construction and application
A prbe-hcr-haat-hfixml, pt-hcr-haat technology, applied in the direction of recombinant DNA technology, introduction of foreign genetic material using a vector, etc., can solve the problem that the amount of gene expression product needs to be further improved, and the efficiency of in vivo mediated integration is not high. problem, to achieve high expression, increase integration efficiency, and improve the effect of expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: The acquisition and purification of pRBE-CMV-FIXml without the CMV promoter vector
[0026] According to pRBE-CMV-FIX sequence (GeneTher.2009;16(5):589-95.JMolBiol.2006;358(1):38-45.), design specific primers (see Table 1-primers), meanwhile, in Restriction sites BglII and NheI were added to the 5' and 3' ends, respectively. Using pRBE-CMV-FIX as a template, the target band was amplified.
[0027] Table 1 - Primers
[0028]
[0029]
[0030] Prepare the PCR reaction system as shown in the table below for cloning pRBE-CMV-FIXml without the CMV promoter vector sequence:
[0031] Element
volume
wxya 2 o
72μl
PCR Buffer
10μl
dNTP
2μl
Mg 2+
8μl
Forward Primer (TEST-Bgl II)
2μl
Reverse Primer (TEST-Nhe I)
2μl
template
3μl (~100ng)
KOD plus neo
1μl
total
100μl
[0032] Carry out the PCR reaction according to the program shown in the ta...
Embodiment 2
[0036] Embodiment 2: "NheI-hAAT-BglII" fragment amplification
[0037] Using human genomic DNA (AmpFlSTR standard DNA, Applied Biosystems, USA) as a template, the NheI-hAAT-BglII DNA fragment was amplified with an upstream primer containing NheI and a downstream primer containing BglII. Prepare the PCR reaction system as shown in the table below for the amplification of the -NheI-hAAT-BglII-fragment:
[0038] Element
volume
wxya 2 o
2μl
Buffer
20μl
[0039] dNTP
2.5μl
Forward Primer (hAAT-F)
1μl
Reverse Primer (hAAT-R)
1μl
template
1μl
HS Taq
25μl
total
50μl
[0040] Carry out the PCR reaction according to the program shown in the table below:
[0041]
[0042] Prepare agarose gel with a concentration of 2%, and conduct constant voltage electrophoresis with a voltage of 120V. Use the AxyPrepDNA Gel Extraction Kit to recover the target DNA fragment.
[...
Embodiment 3
[0044] Embodiment 3: "SpeI-HCR-SpeI" fragment amplification
[0045] Using plasmid human genomic DNA (AmpFlSTR standard DNA, Applied Biosystems, USA) as a template, the SpeI-HCR-SpeI DNA fragment was amplified with SpeI-containing upstream primers and SpeI-containing downstream primers. Prepare the PCR reaction system as shown in the table below for the amplification of the -SpeI-HCR-SpeI-fragment:
[0046] Element
volume
wxya 2 o
14.3μl
Buffer
2μl
dNTP
0.4μl
Forward Primer (F-Spe I)
0.4μl
Reverse Primer (R-Spe I)
0.4μl
Mg +
1.6μl
template
0.5μl
Taq
0.4μl
total
20μl
[0047] Carry out the PCR reaction according to the program shown in the table below:
[0048]
[0049]Prepare agarose gel with a concentration of 2%, and conduct constant voltage electrophoresis with a voltage of 120V. Use the AxyPrepDNA Gel Extraction Kit to recover the target DNA frag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com