Kluyveromyces lactis eukaryotic expression method of streptomyces murinus AMP deaminase gene
A technology of Streptomyces griseus and Kluyveromyces, applied in the field of molecular biology, to achieve high and stable enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Cloning of embodiment 1 Streptomyces griseus AMP deaminase gene
[0027] Using the genome of Streptomyces griseis MCCC1A01641 as a template, the primer pair AMPDF3 / AMPDR3 was designed to specifically amplify the AMP deaminase gene of Streptomyces griseris , and XhoI restriction sites and BglII restriction sites were added at both ends of the primer pair. The AMPDF3 / AMPDR3 sequence is as follows: AMPDF3: 5'-GTT CTCGAG GCGCCGCCGCCCCGGCAG-3' (SEQ ID NO: 1); AMPDR3: 5'-GA AGATCT TCACCCCCGGGCGTGCGCCC-3' (SEQ ID NO: 2);
[0028] Purify and recover the PCR product.
Embodiment 2
[0029] Example 2 Construction of recombinant expression vector pKLAC1-AMPD
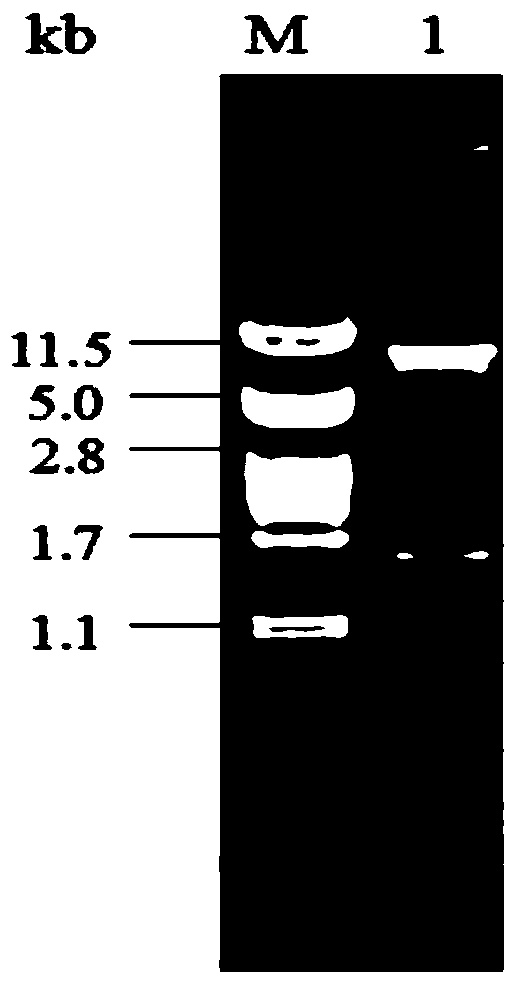
[0030] The vector pKLAC1 and the PCR product obtained in Example 1 were double-digested with XhoI and BglII respectively, recovered and purified by gel cutting, the digested product was ligated overnight at 16°C, the ligated product was transformed into E.coli JM109, and the pKLAC1-AMPD-JM109 strain plasmid was extracted. The pKLAC1-AMPD-JM109 positive strain was screened out by XhoI and BglII double enzyme digestion. Recombinant expression vector pKLAC1-AMPD enzyme digestion verification electrophoresis picture is as follows figure 1 shown.
Embodiment 3
[0031] Example 3 Electrotransformation of linearized plasmid pKLAC1-AMPD to GG799 and screening of transformants
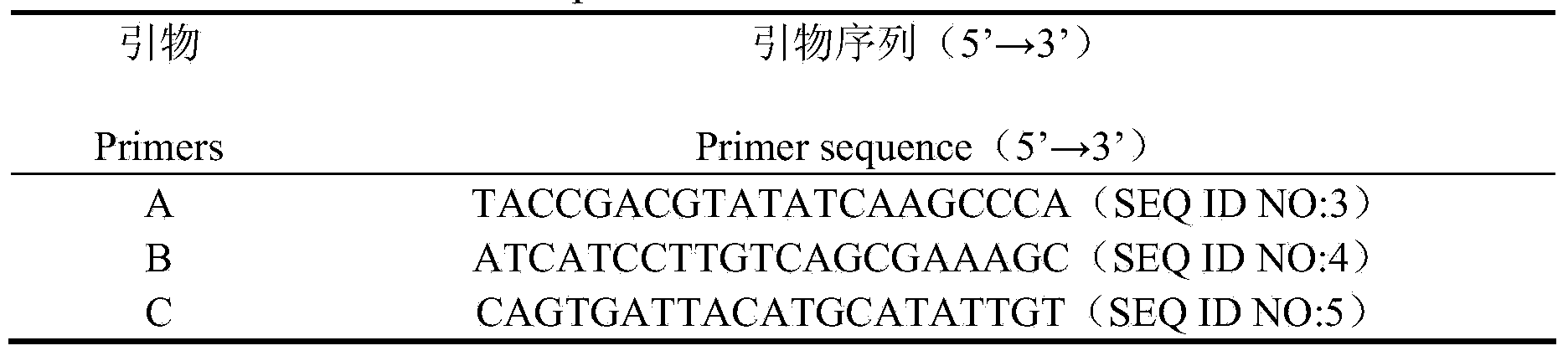
[0032] Extract the pKLAC1-AMPD-JM109 positive strain plasmid obtained in Example 2, linearize it with BstXI, and electrotransform Kluyveromyces lactis GG799, spread it on a YCB plate, pick a single colony for detection after 3 to 5 days, and extract the recombinant chromosome genome by the helicase method , using specific primers AMPDF3 / AMPDR3 to carry out PCR verification to obtain positive clone transformants; then use the universal primers that come with the pKLAC1 vector to perform PCR to screen out multi-copy transformants. The sequences of the universal primers are shown in Table 1:
[0033] Table 1 General primer sequence of pKLAC1 vector
[0034]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com