Streptomycete generating 5-keto-milbemycins and method for producing 5-keto-milbemycins
A technology of mirbemycin and streptomyces, which is applied in the field of producing 5-keto mirbemycin and can solve the problems of shortening the production process of mirbemycin and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0140] Embodiment 1: the extraction of Streptomyces total DNA
[0141] Inoculate 50 μL of Streptomyces spore suspension in cryopreserved tubes into 30 mL of TSB medium (purchased from Bacto Tryptic Soy Broth.BD), culture at 28°C and 220 rpm for 48 hours, centrifuge at 4000 rpm for 10 minutes in a 50 mL centrifuge tube, and remove the supernatant. The precipitate was washed twice with 30 mL of sucrose-Tris buffer (in which the mass percentage of sucrose was 10.3%, the molar-volume concentration of Tris-HCl was 10 mM, and the pH value was 8.0), and then suspended in 5 mL of sucrose-Tris buffer. Add 20 μL of lysozyme solution with a mass-volume solubility of 100 mg / mL, and bathe in water at 37° C. for 2 h. Add 500 μL of 10% SDS solution by mass percentage, and gently invert until it is basically clear. Add phenol-chloroform-isoamyl alcohol (the volume ratio of phenol-chloroform-isoamyl alcohol is 25:24:1 (pH value is 8.0)) solution 5mL, after gentle inversion several times, cent...
Embodiment 2
[0142] Embodiment 2: The construction of the recombinant vector that is used for knocking out Streptomyces milF gene
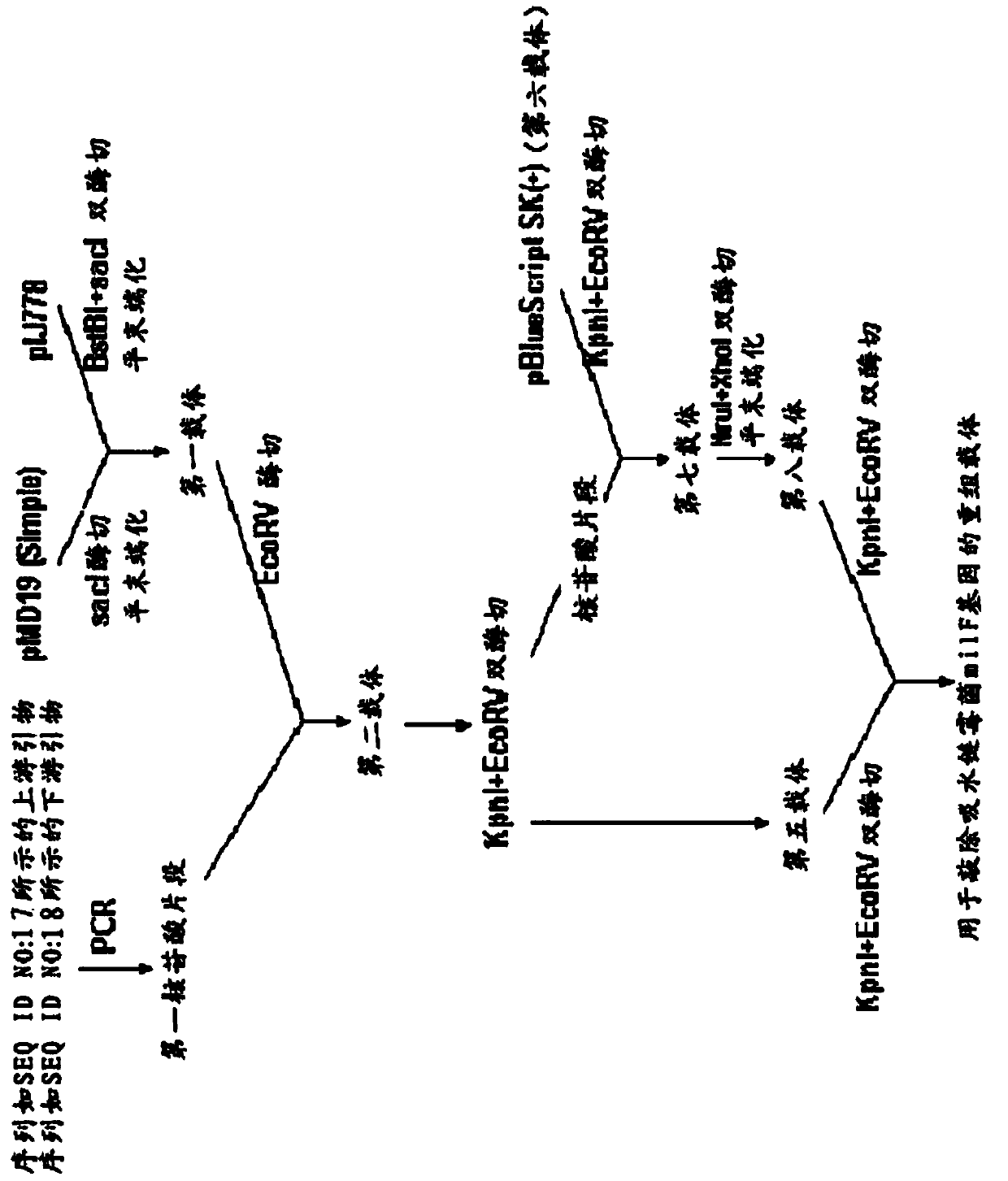
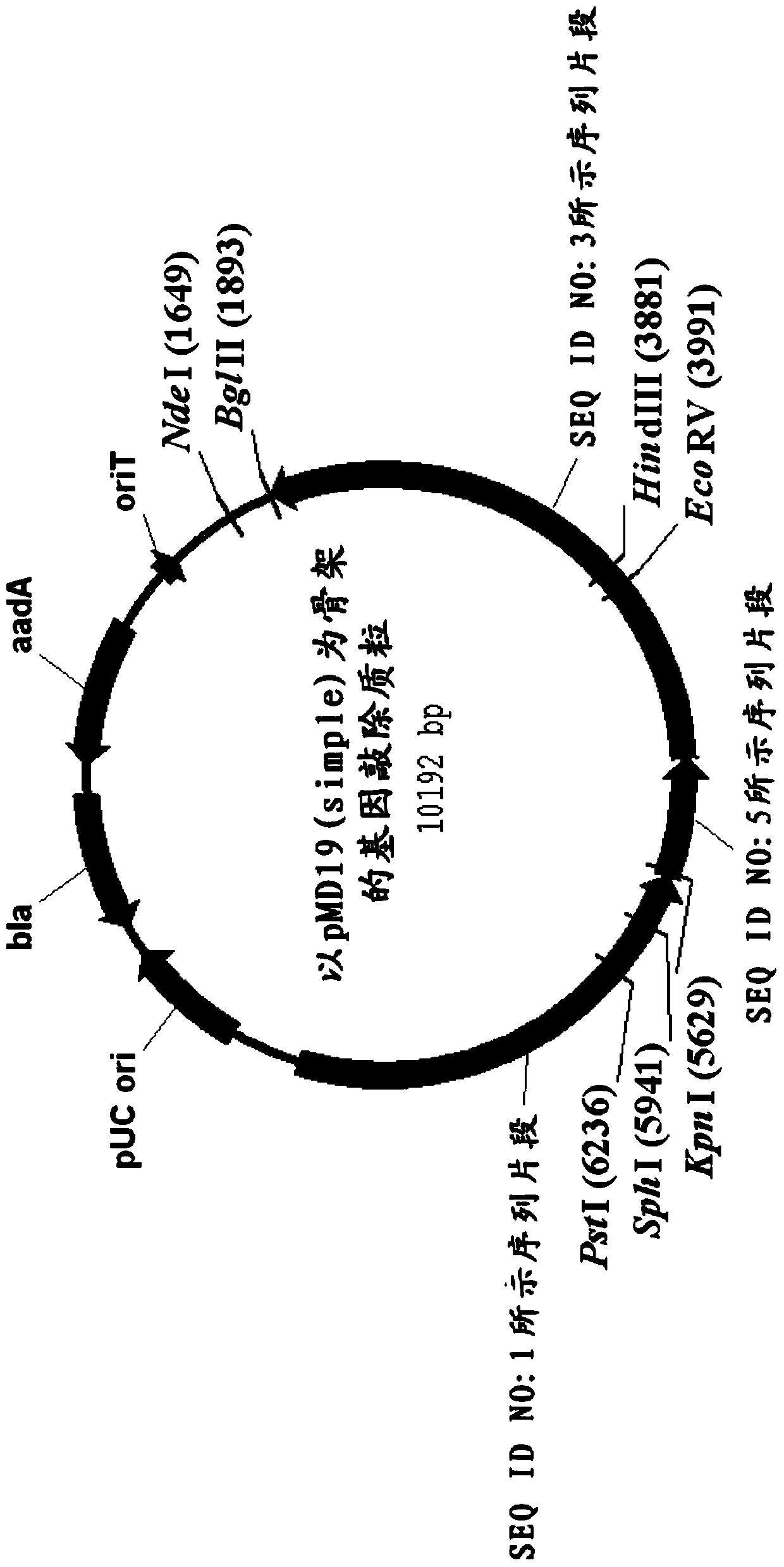
[0143] With pMD19 (Simple) as the backbone, its construction process is as follows figure 2 As shown, specifically:
[0144] a): using the Streptomyces total DNA obtained in Example 1 as a template, using an upstream primer with a nucleotide sequence as shown in SEQ ID NO: 17, and a downstream primer with a nucleotide sequence as shown in SEQ ID NO: 18, through amplifying to obtain the first nucleotide fragment;
[0145] 1. Preparation of PCR amplification reaction solution, in which PrimeSTAR kit was purchased from TaKaRa:
[0146]
[0147] 2. Divide into 2 tubes for PCR reaction, the procedure is:
[0148]
[0149] After agarose gel electrophoresis, the PCR product was recovered with a DNA gel recovery kit (purchased from Shanghai Huasun Biotechnology Co., Ltd.) to obtain the first nucleotide fragment, whose sequence is shown in SEQ ID NO:36.
[0...
Embodiment 3
[0159] Embodiment 3: The construction of the recombinant vector that is used for knocking out Streptomyces milF gene
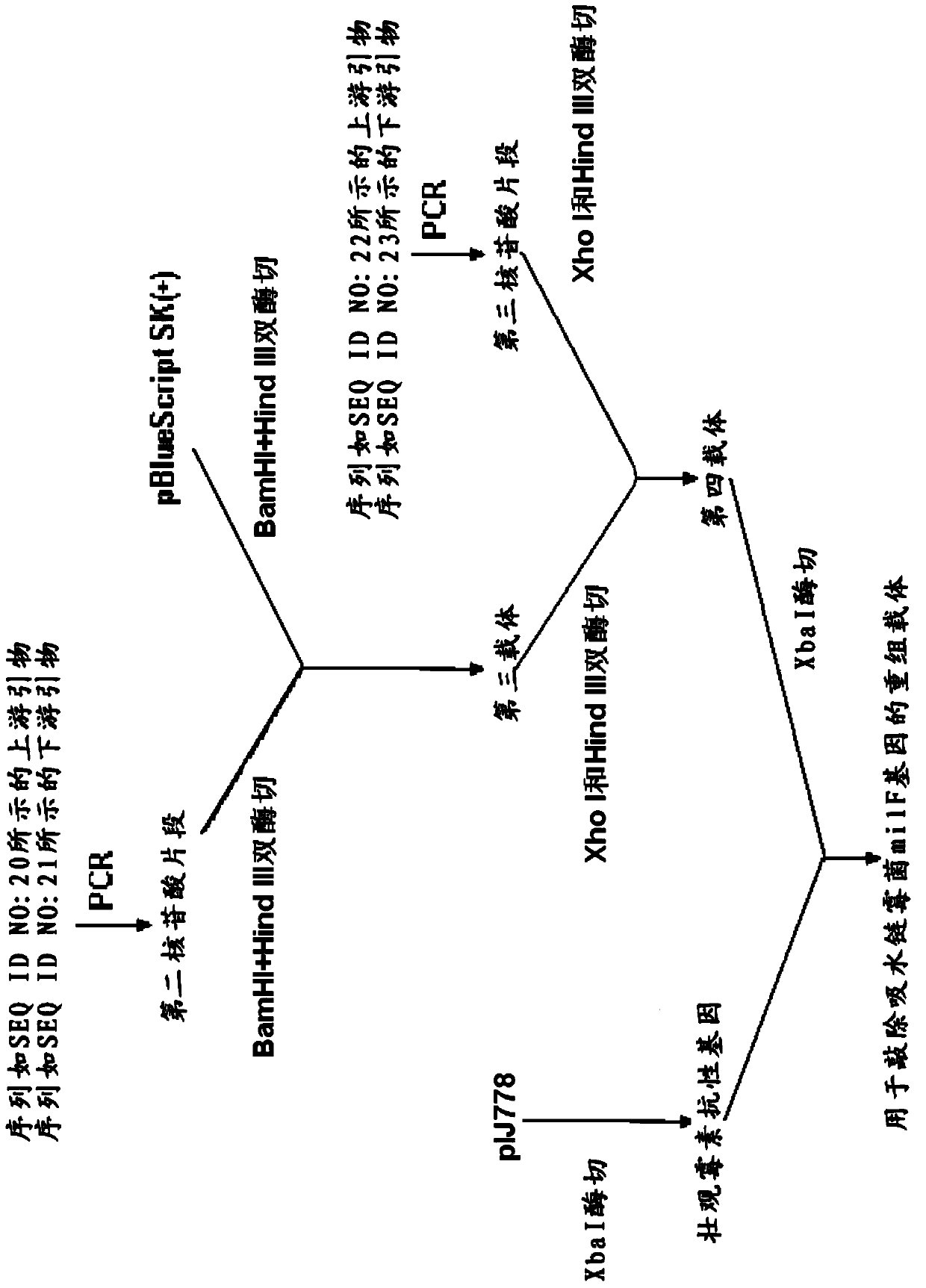
[0160] With pBlueScript SK(+) as the skeleton, its construction process is as follows Figure 4 As shown, specifically:
[0161] a) Using the total Streptomyces DNA as a template, using an upstream primer with a nucleotide sequence as shown in SEQ ID NO: 20, and a downstream primer with a nucleotide sequence as shown in SEQ ID NO: 21, PCR amplifies to obtain SEQ The second nucleotide fragment of the nucleotide sequence shown in ID NO:24:
[0162] 1. Preparation of PCR amplification reaction solution, in which PrimeSTAR kit was purchased from TaKaRa:
[0163]
[0164] 2. Divide into 2 tubes for PCR reaction, the procedure is:
[0165]
[0166] After agarose gel electrophoresis, the PCR product was recovered with a DNA gel recovery kit (purchased from Shanghai Huasun Biotechnology Co., Ltd.) to obtain a second nucleotide fragment, the nucleotide sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com