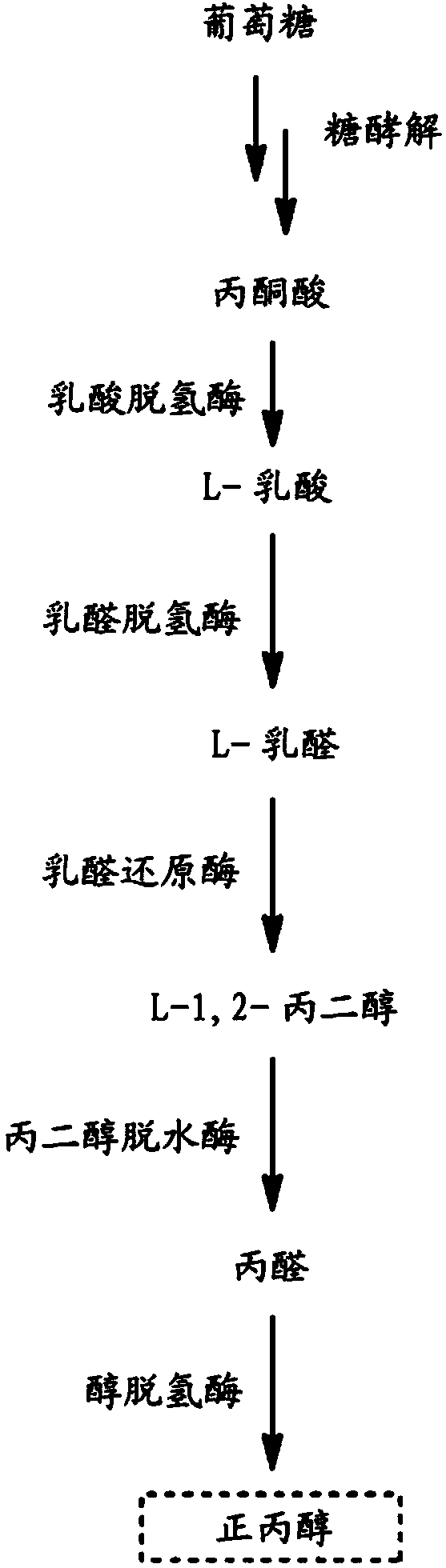
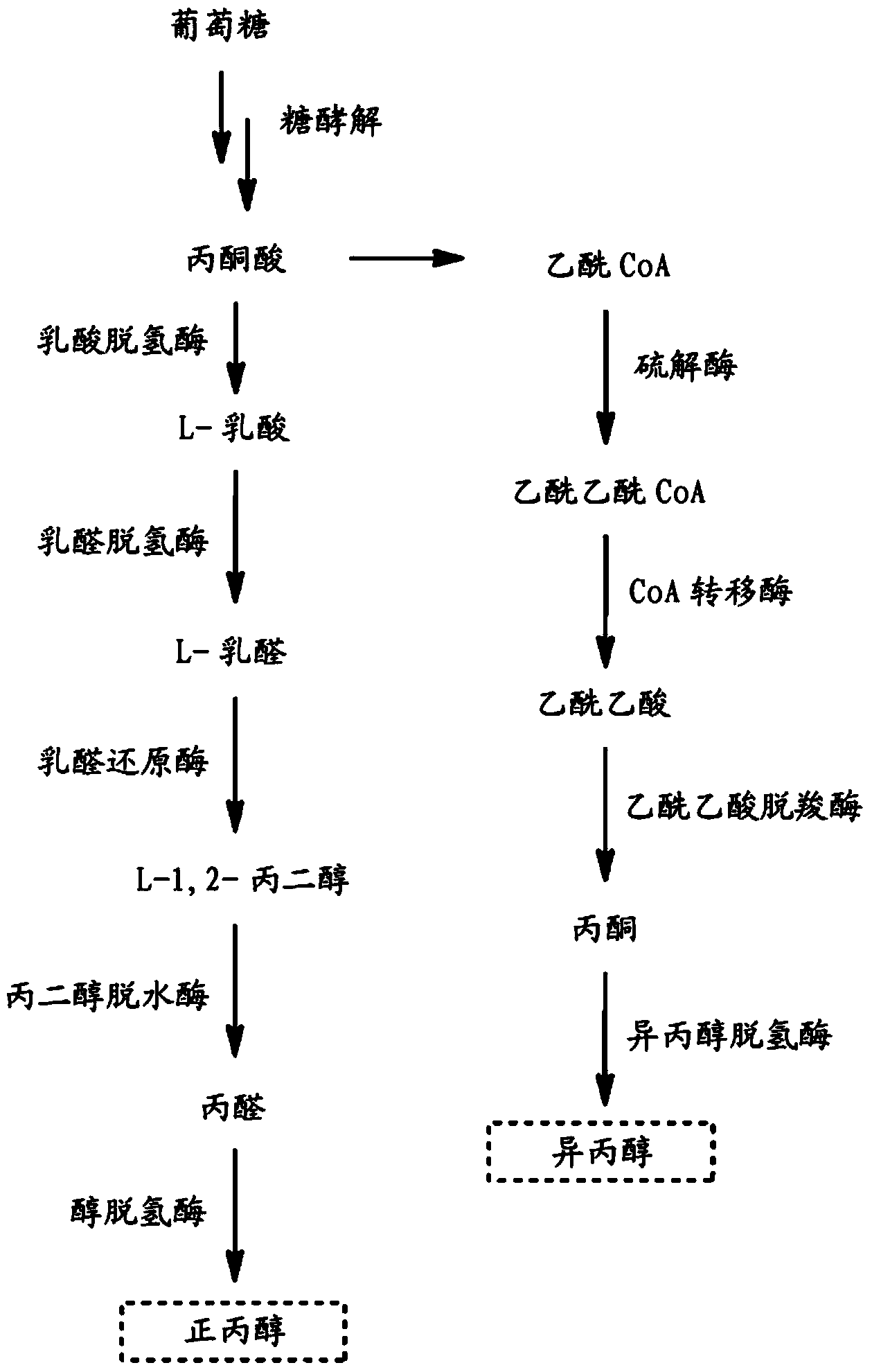
Microorganisms for n-propanol production
A technology of n-propanol and recombinant host cells, applied in fermentation, bacteria, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0436] Example 1: Cloning of Clostridium acetobutylicum Thiolase Gene and Construction of Vector pTRGU51
[0437] The 1176 bp coding sequence (CDS) of the thiolase gene identified in Clostridium acetobutylicum was optimized for expression in E. coli and synthetically constructed into pTRGU51. A DNA fragment containing a codon-optimized CDS was designed to have a ribosome binding site (RBS, sequence 5'-GAAGGAGATATACC-3') immediately before the start codon. The resulting sequence was then submitted to and synthesized by Geneart AG (Regenburg, Germany) and delivered into a pMA backbone vector containing the β-lactamase encoding gene blaTEM-1. When synthesized, the CDS and RBS fragments are flanked by restriction sites to facilitate subsequent cloning steps. The entire synthetic fragment cloned into the pMA vector was EcoRI-RBS-CDS-STOP-BamHI-XbaI resulting in pTRGU51.
[0438] The wild-type nucleotide sequence (WT) and deduced amino acid sequence of the Clostridium acetobutylic...
Embodiment 2
[0439] Embodiment 2: Bacillus subtilis succinyl CoA: the cloning of acetoacetate transferase gene and the construction of vector pTRGU58 and pTRGU59
[0440] The 699bp coding sequence (CDS) of the scoA subunit of the subtilis succinyl CoA: acetoacetate transferase gene and the 648bp coding sequence of the scoB subunit of the subtilis succinyl CoA: acetoacetate transferase gene are optimized for use in the large intestine Bacteria and were synthetically constructed into pTRGU58 and pTRGU59, respectively. Each DNA fragment containing a codon-optimized CDS was designed with a ribosome binding site and synthesized by Geneart AG (Regenburg, Germany) as described in Example 1 with the modified restriction sites shown below point. The entire synthetic fragment containing scoA cloned into the pMA vector was EcoRI-BamHI-RBS-scoA-STOP-NotI-XbaI resulting in pTRGU58. The entire synthetic fragment containing scoB cloned into the pMA vector was EcoRI–NotI–RBS–scoB–STOP–HindIII–XbaI, and ...
Embodiment 4
[0448] Example 4: Cloning of Clostridium beijerinckii acetoacetate decarboxylase gene and construction of vector pTRGU62
[0449] The 738 bp coding sequence (CDS) of the acetoacetate decarboxylase gene of C. beijerinckii was optimized for expression in E. coli and synthetically constructed into pTRGU62. A DNA fragment containing a codon-optimized CDS was designed with a ribosome binding site and synthesized by Geneart AG as described in Example 1 with the modified restriction sites shown below. The entire synthetic fragment cloned into the pMA vector was EcoRI-HindIII-RBS-CDS-STOP-AscI-XbaI resulting in pTRGU62.
[0450] The wild-type nucleotide sequence (WT) and deduced amino acid sequence of C. beijerinckii acetoacetate decarboxylase correspond to SEQ ID NO: 47 and 48, respectively. The coding sequence is 741 bp, including the stop codon, and the encoded predicted protein is 246 amino acids. No signal peptide was predicted in this sequence using the SignalP program (Nielse...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com