Database searching method and database searching system for open type protein identification
A protein identification and database technology, applied in the field of bioinformatics, to achieve the effect of improving resolution, facilitating retrieval, and increasing scale
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0062] The present invention will be described in detail below in conjunction with the accompanying drawings and specific embodiments, but not as a limitation of the present invention.
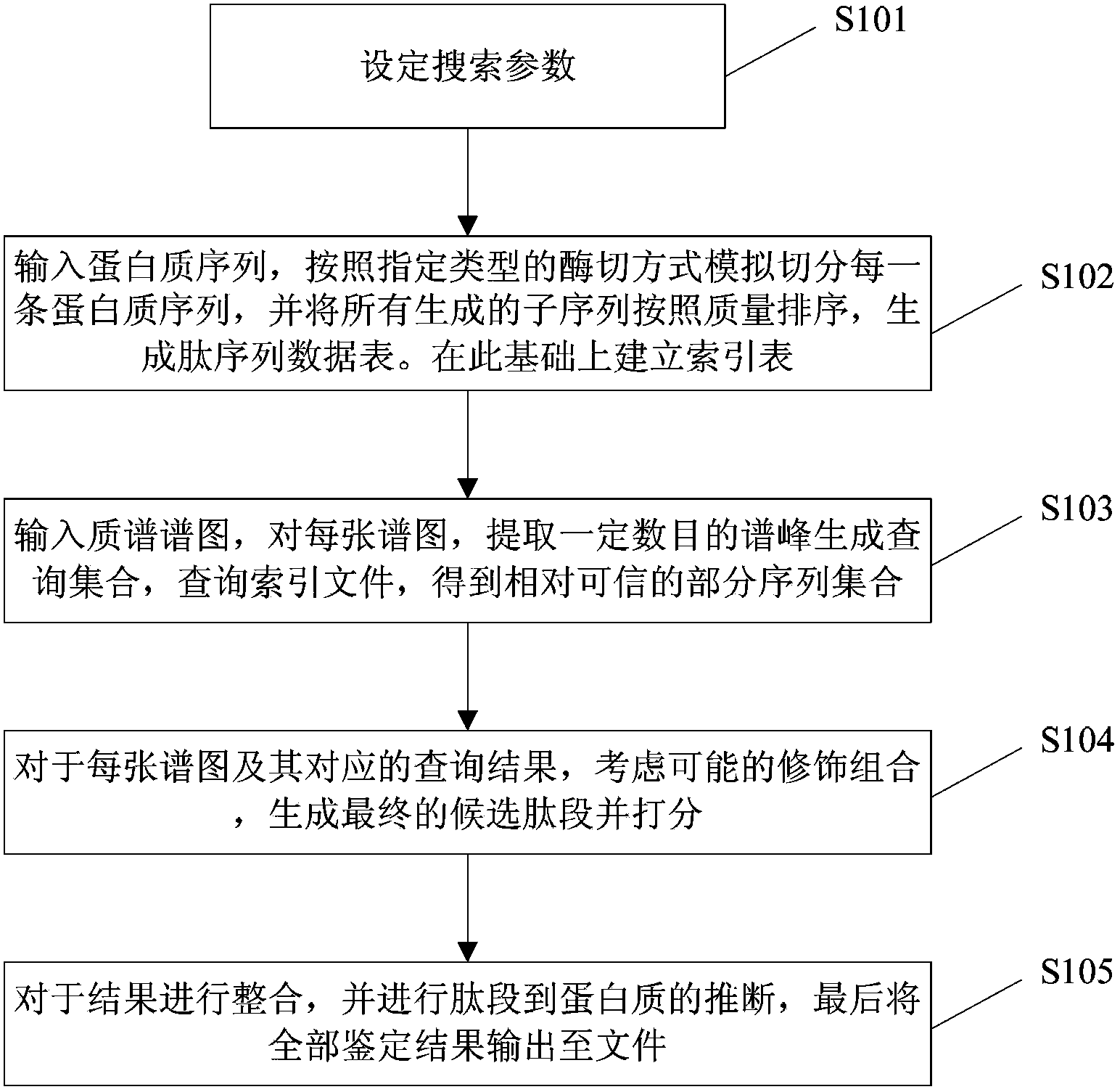
[0063] Such as figure 1 Shown is a flow chart of the search method for the open protein identification database of the present invention. The specific steps of the process are as follows:
[0064] Step 101, setting necessary search parameters.
[0065] Step 102, inputting the protein sequence, slicing each protein sequence according to the specified type of enzyme cutting method, sorting all generated subsequences according to quality, and generating a peptide sequence data table. Index files are built on this basis.
[0066] Step 103, input the mass spectrogram, extract a certain number of spectral peaks from each mass spectrogram to generate a query set, and then query the index file described in step 101 to obtain the query result. The query result is a sequence fragment, that is, a rel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com