A method for detecting single nucleotide polymorphism of goat stat3 gene and its application
A gene and goat technology, applied in the field of detection of goat STAT3 gene single nucleotide polymorphism, can solve problems such as blank and lack of functional research of gene loci
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
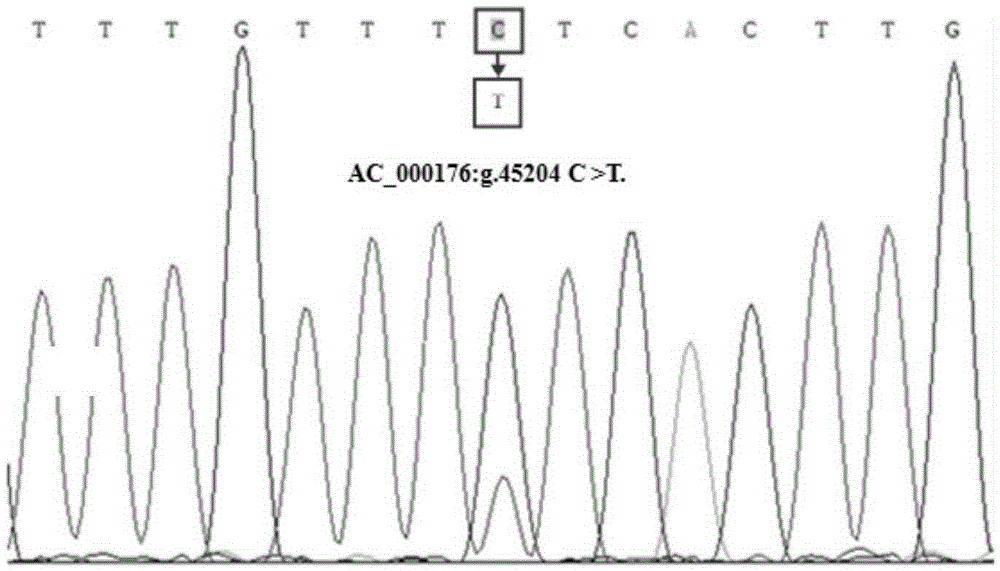
[0066] Embodiment 1, goat STAT3 gene contains the design of 45204, 62058 and 62230 PCR primers
[0067] Since cattle and sheep are both ruminants and highly homologous, and the sequence of the goat genome has not been released so far, the sequence of the bovine STAT3 gene can only be searched on NCBI, and Primer5. 2, 10 and 11 intronic PCR primers for SNP sites, the primer sequences are as follows:
[0068] P1: Upstream primer: 5'-TGTGTGAGTGTGTAGGTTTCAGAAT-3'(25bp),
[0069] Downstream primer: 5'-AGATAGGTTATTAATTAATCAACT A-3'(26bp);
[0070] P2: Upstream primer: 5'-AAATCCAGCCTGAGGGAGAATTCCT-3'(25bp),
[0071] Downstream primer: 5'-GAGTCTCTTTGAAAGTCCACTTTG A-3'(26bp);
[0072] P3: Upstream primer: 5'-TGGGAAAGATACTTGCTG-3'(18bp),
[0073] Downstream primer: 5'-GACCTGAATCACAGGAGGAAAAGA C-3'(26bp)
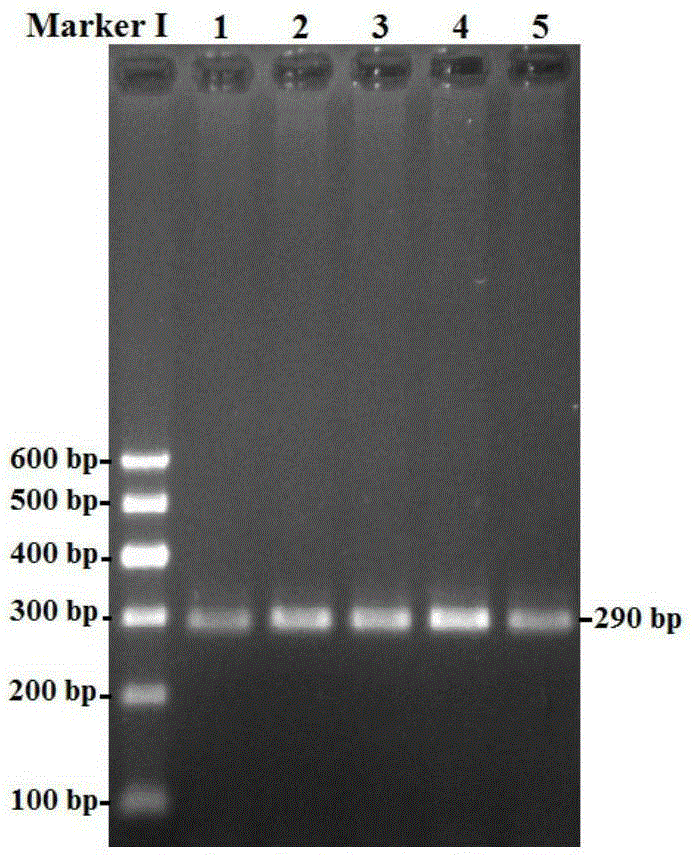
[0074] Using the above primer pair P1 to amplify the goat genome, it is possible to amplify a 290bp fragment including the 44941nt-45230nt in the second intron region of the...
Embodiment 2
[0083] Embodiment 2, carry out PCR amplification STAT3 gene fragment of goat to be tested with primer P
[0084] 1. Collection of goat samples
[0085] The animals used in the experiment were a total of 585 samples of two breeds, of which
[0086] 1) 220 samples of Hainan Black Goat (HNBG) were collected from the Hainan Black Goat Breeding Farm in Zhanzhou, Hainan Province. Random sampling method was used to collect individual ear tissue samples from the Hainan Black Goat Breeding Farm in Danzhou City, Hainan Province. These samples were stored in 70% ethanol, brought back to the laboratory in an ice box and stored at -80°C.
[0087] 2) A total of 365 samples of Xinong Sanen dairy goat (XNSN) were collected from the Saaneng dairy goat breeding farm in Qianyang County, Shaanxi Province. Blood samples from individuals in the breeding field were collected randomly, anticoagulated with ACD, brought back to the laboratory at low temperature in an ice box, and stored at -80°C.
...
Embodiment 3
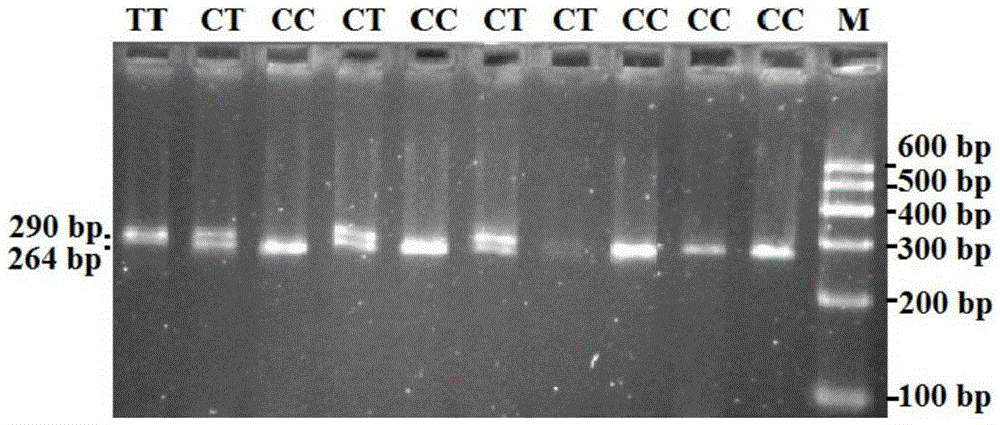
[0143] Different STAT3 gene fragments of embodiment 3, DdeI, RsaI and TaqI digestion digestion PCR amplification
[0144] 1. DdeI, RsaI and TaqI digestion reaction digestion system
[0145] DdeI, RsaI and TaqI digestion reaction systems are all 10 μL system, including 4.0 μL of PCR product, 0.2 μL of endonuclease, 1.0 μL of 10× buffer, 1.0 μL of 0.1% BSA and 3.8 μL of deionized water.
[0146] 2. Enzyme digestion conditions:
[0147] DdeI, RsaI enzyme digestion conditions are 37 ℃ overnight (12 ~ 16h), TaqI enzyme digestion conditions are placed in a constant temperature water bath at 65 ℃ overnight (12 ~ 16h).
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com