A Label-free Quantitative Method for Proteome Combining Secondary Mass Spectrometry and Machine Learning Algorithms
A secondary mass spectrometry and machine learning technology, applied in the quantitative field of proteomics, to achieve the effect of reliable identification results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
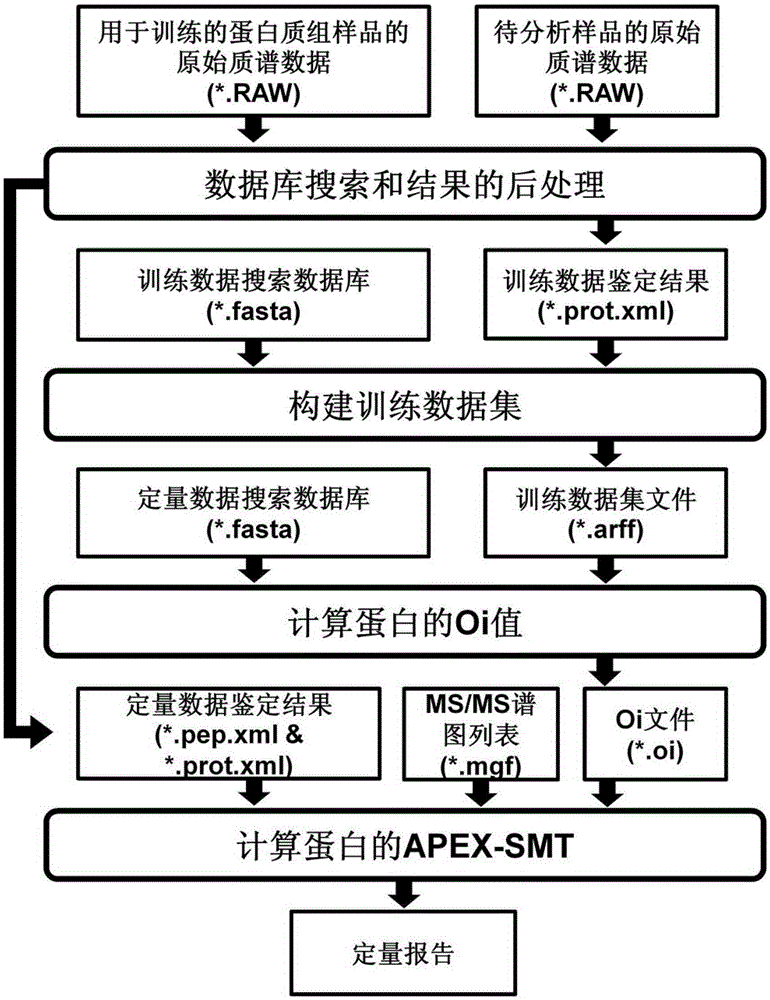
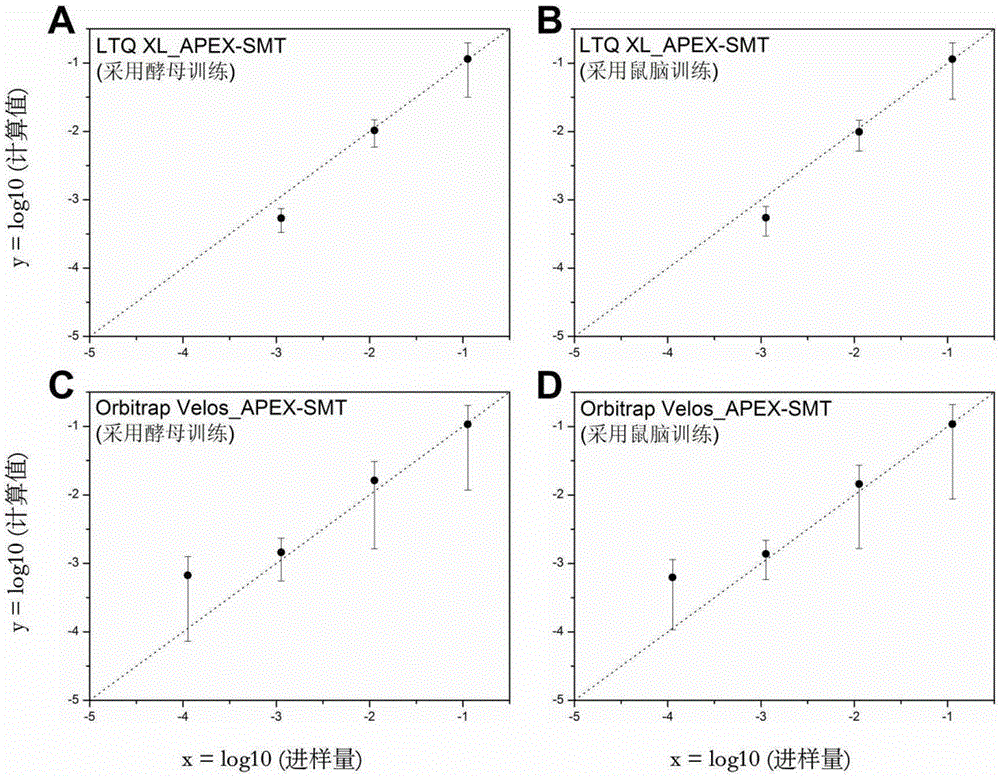
[0020] 1. The trypsin digests of proteins extracted from yeast and mouse brain were used as the data source of the training data set, and they were run 5 times on the one-dimensional nano-RPLC-MS / MS system respectively. Mass spectrometers are LTQXL and OrbitrapVelos from Thermo Company. The UPS2 standard protein mixture purchased from Sigma was used as the quantitative data set to test the effect of the method. The UPS2 standard protein mixture is composed of 48 human-derived standard proteins, the concentration of which spans 6 orders of magnitude, and there are 8 proteins with different properties in each order of magnitude. The tryptic digest of UPS2 was run 5 times on the same system. The absolute amount of UPS2 on the column spans 6 orders of magnitude from 5 amol to 500 fmol.
[0021]The original data RAW file was converted into mgf format with the msconvert.exe component in TPP (version4.6), and then the mgf file was searched using the Mascot (version2.3.02) database ...
Embodiment 2
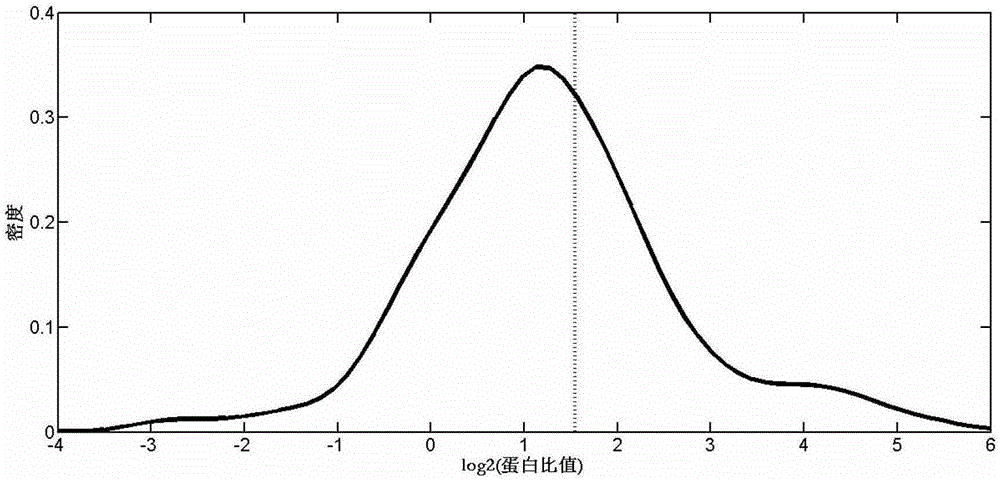
[0028] 1. The relatively quantitative test data set comes from the Clinical Proteomic Technology Assessment for Cancer (CPTAC), from http: / / www.proteomecommons.org / Downloaded from the website (hash:NGX3cBUAZXSWvc+6XFNIdVhpLPJTO87lzAxUQmwwR2KHUwWDrdFwV1dso3bvxf7HeXZ4C / juqwEUIz4boC9H3HcLrxEAAAAAAAmDw==), the name of the dataset is Study6OrbitrapO86. This data set contains 5 samples A-E, each sample contains an equal amount of yeast extract protein (60ng / μL), and contains 0.24, 0.74, 2.2, 6.7, 20fmol / μL of UPS1 standard protein mixture, so that every two The fold change of UPS1 protein in adjacent samples was 3-fold while the content of yeast protein was unchanged. UPS1 is similar to UPS2 except that the 48 standard proteins are mixed in equimolar proportions. Each sample was injected in triplicate on the OrbitrapXL mass spectrometer. Since the UPS1 protein content in samples A and B is extremely low, the yeast protein in them was used as the data source of the training data ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com