PCR kit used for detecting CGC replication number and AGG insert information of fragile X syndrome
An X chromosome and information insertion technology, applied in the field of PCR kits, can solve the problems of difficult operation, long detection time, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
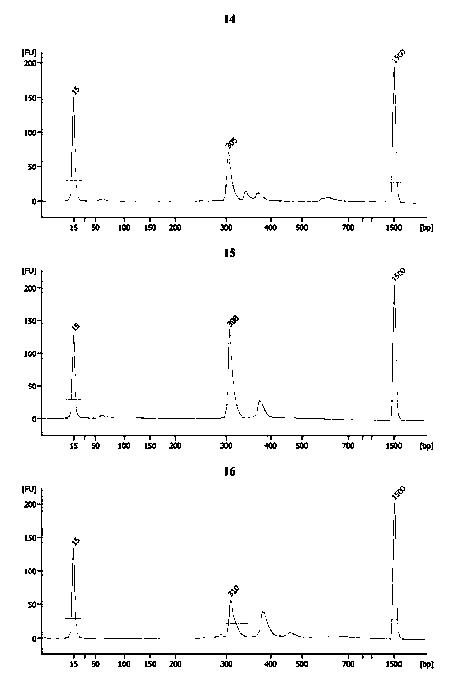
[0035] Example 1 Preparation of PCR kit for CGG repeat number and AGG insertion information of fragile X syndrome
[0036] 1. Primer design
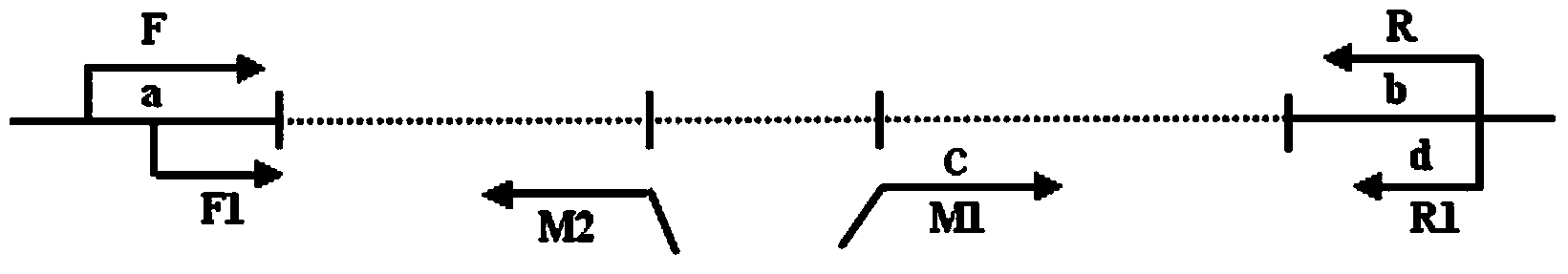
[0037]A pair of primers F and R flanking the CGG repeat region are provided for amplifying the CGG repeat region; two different pairs of primers are provided in addition, including the first primer containing the search probe and the corresponding annealing primer located on the flank of the CGG repeat region; one of One pair is detection primers M1 and R1, and the other pair is verification primers F1 and M2. Two pairs of different primers each contain a first primer M1 and M2 as a search probe, wherein the first primer M1 of the detection primer M1 and R1 is used as an upstream primer, and the first primer M2 of the verification primer M2 and F1 is used as a downstream primer. primers. M1 includes CGG, GCG, GGC repeats and AGG, GAG, GGA repeats, M2 includes GCC, CGC, CCG repeats and TCC, CTC, CCT repeats. M1 and M2 consist of a sequ...
Embodiment 2
[0043] Example 2 CGG repeat number accurate quantification
[0044] 1) Use a commercially available genomic DNA extraction kit to extract human peripheral blood DNA, the concentration of which is 100 ng / μl, the 260 / 230 value is greater than 2.0, and the 260 / 280 value is greater than 1.8.
[0045] 2) Prepare PCR reaction system: prepare 50 μl PCR reaction system in a 200ul thin-walled PCR test tube,
[0046] The PCR amplification system is as follows:
[0047]
[0048] Wherein, the primers are SEQ ID No: 2 and SEQ ID No: 4.
[0049] Composition of PCR enhancer: betaine (2.5mol / L) 5 μl, GCEnhancer 5 μl and DMSO 4 μl.
[0050] PCR reaction program: 95°C pre-denaturation for 8 min, followed by 30 cycles: denaturation at 95°C for 50 s, annealing at 65°C for 45 s, extension at 72°C for 1 min, and final extension at 72°C for 10 min.
[0051] 3) After the reaction, use 2% agarose gel to load the sample, electrophoresis under the electric field condition of 10V / cm for 45min, and ...
Embodiment 3A
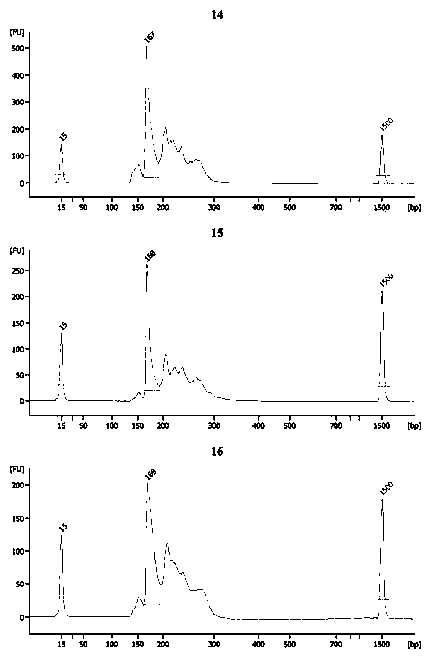
[0057] Example 3 AGG Insertion Information Accurate Quantification
[0058] 1) Use a commercially available genomic DNA extraction kit to extract human peripheral blood DNA, the concentration of which is 100 ng / μl, the 260 / 230 value is greater than 2.0, and the 260 / 280 value is greater than 1.8.
[0059] 2) Prepare PCR reaction system: Prepare 50 μl PCR reaction system in a 200ul thin-walled PCR tube, and the PCR amplification system is as follows:
[0060]
[0061]
[0062] Wherein, the primers are SEQ ID No: 5 and SEQ ID No: 11.
[0063] Composition of PCR enhancer: betaine (2.5mol / L) 4 μl and GCEnhancer 4 μl.
[0064] PCR reaction program: 95°C pre-denaturation for 8 min, followed by 30 cycles: denaturation at 95°C for 50 s, annealing at 68°C for 45 s, extension at 72°C for 1 min, and final extension at 72°C for 10 min.
[0065] 3) After the reaction, use 2% agarose gel to load the sample, electrophoresis under the electric field condition of 10V / cm for 45min, and u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com