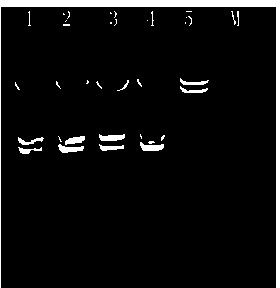Panx3 gene overexpression red fluorescence lentiviral vector and building method and application thereof
A lentiviral vector and gene overexpression technology, applied in the field of genetic engineering, can solve the problem of mutual interference between overexpression vectors and functional research, and achieve the effect of high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Construction of pCDNA3.1(+)-Panx3
[0040] Total RNA was extracted from human dental pulp cells, using the reverse-transcribed cDNA as a template, primers were designed according to the sequence of the Panx3 gene cDNA, and PCR amplification was performed with primers 1 and 2 as primers.
[0041] Primer 1: 5'-CCC AAGCTT ATGTCACTTGCACACACAG-3',
[0042] Primer 2: 5'-CGC AAGCTT TTTGACTCTTCGGCTCCA-3' (underlined bases are HindIII recognition sites).
[0043] The PCR reaction conditions were: 94°C for 3min, followed by 35 cycles of 94°C for 30s, 58°C for 40s, 72°C for 1min, and finally 72°C for 10min.
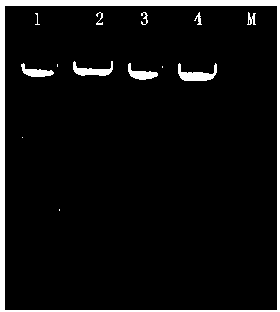
[0044]A linear DNA amplification product Panx3 (1396bp) can be obtained by PCR, with HindIII restriction sites at both ends. The Panx3 fragment and the pCDNA3.1(+) plasmid are simultaneously digested with HindIII, and recovered by the gel recovery kit (Kangwei Century Company), T4 DNA ligase for two-fragment ligation, and transform E.coli DH5α competent cells ...
Embodiment 2
[0045] Example 2 Construction of pCDNA3.1(+)-Panx3-RFP
[0046] The Thy-Brainbrow-1.0H (Addgene China agency company, Plasmid 18724) vector was used as a template, and the RFP red fluorescent protein fragment was amplified by PCR with primers 3 and 4 as primers.
[0047] Primer 3: 5'-TCC CCCGGG AGACCCAAGCTGGCTAGCG-3', recognized as dot by SmaI,
[0048] Primer 4: 5'-CCC TTCGAA CTGGCAACTAGAAGGCACAGTCG-3', BstBI recognition site.
[0049] The PCR reaction conditions were: 94°C for 3min, followed by 33 cycles of 94°C for 30s, 58°C for 30s, 72°C for 1min, and finally 72°C for 10min.
[0050] A linear DNA amplification product RFP (805bp) was obtained by PCR. One end contained a SmaI restriction site, and the other end contained a BstBI restriction site. SmaI and BstBI endonucleases were used to simultaneously digest the RFP fragment and pCDNA3.1(+)- The Panx3 recombinant plasmid was recovered by gel recovery kit (Kangwei Century Company), and the two fragments were connected...
Embodiment 3
[0051] Example 3 Construction of pCDNA3.1(+)-Panx3-CMV-RFP
[0052] Using pCDNA3.1(+) as a template, primers 5 and 6 were used to amplify the CMV promoter fragment by PCR,
[0053] Primer 5: 5'-CGC GGATCC GCTTGACCGACAATTGCAT-3', BamHI recognition site,
[0054] Primer 6: 5'-CGC GGATCC ATTTCGATAAGCCAGTAAGCAG-3', BamHI recognition site.
[0055] The PCR reaction conditions were: 94°C for 3min, followed by 33 cycles of 94°C for 30s, 58°C for 30s, 72°C for 1min, and finally 72°C for 10min.
[0056] A linear DNA amplification product CMV fragment (732bp) was obtained by PCR, with BamHI restriction sites at both ends, recovered from the gel extraction kit (Kangwei Century Company), and the fragment and pCDNA3.1( +)-Panx3-RFP recombinant plasmid, use T4 DNA ligase to connect the two fragments, and transform E.coli DH5α competent cells. The kit extracts the pCDNA3.1(+)-Panx3-CMV-RFP recombinant plasmid, digests it with restriction endonuclease NcoI-HF, and the digested product ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com