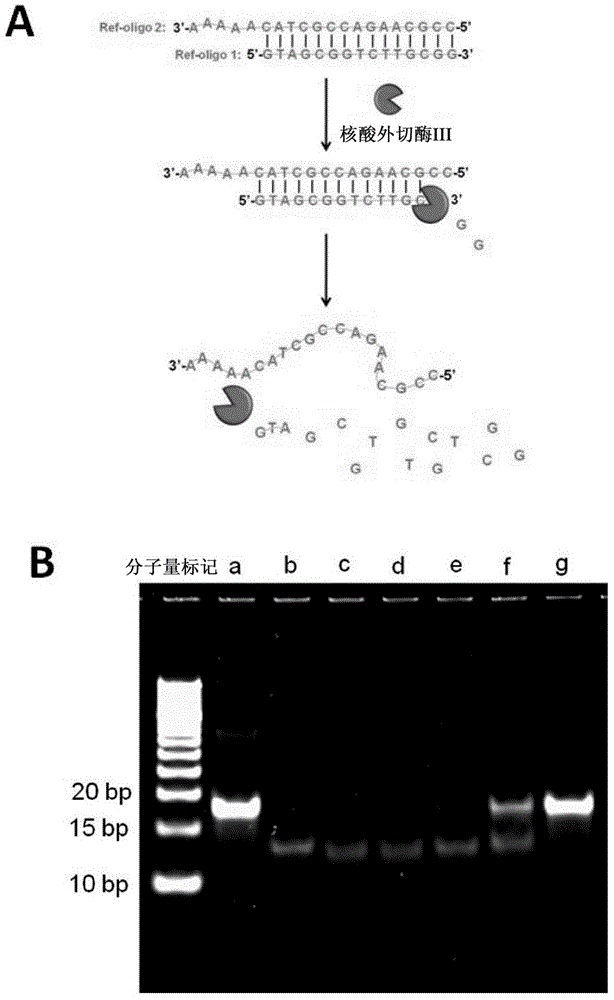
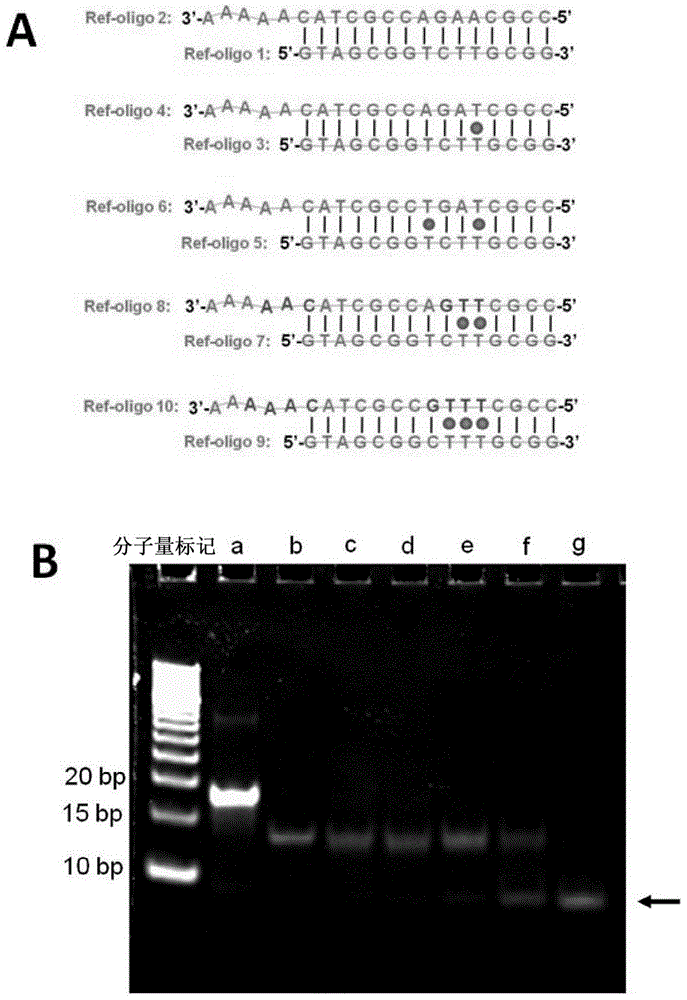
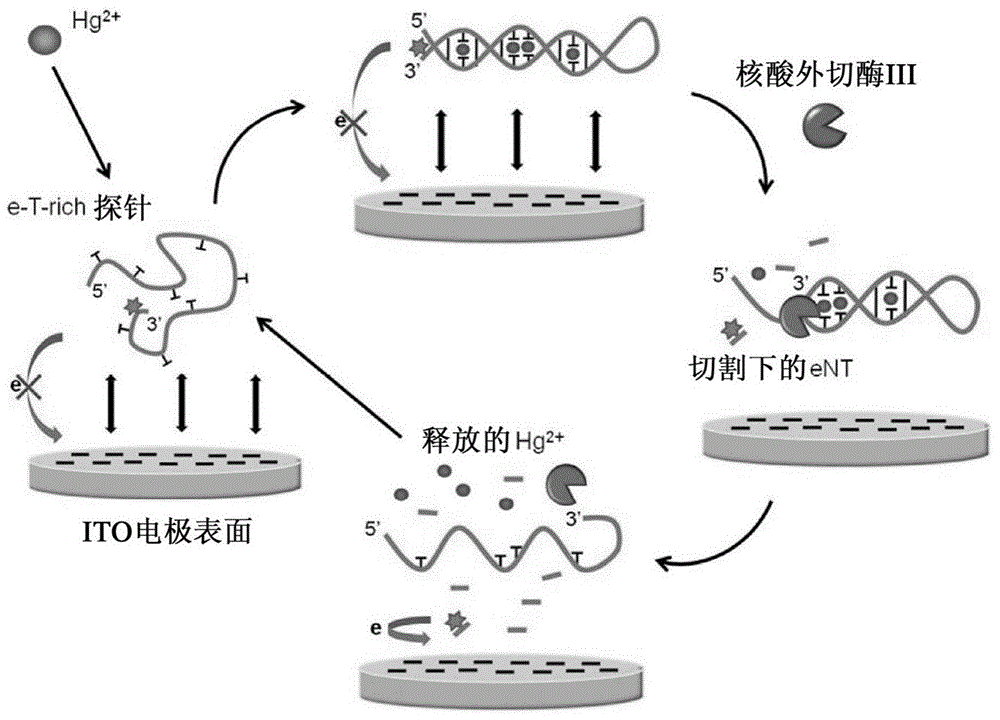
Method For Detecting Mercury Ions In Solution, And Kit
A kit and mercury ion technology, applied in the fields of biochemistry and electrochemistry, can solve the problems of complex fluorescence instruments, which are not suitable for on-site detection, etc., and achieve the effects of improving detection sensitivity, rapid detection, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0112] In 1×NEBuffer1 buffer, e-T-rich single-stranded DNA was mixed with triplicate 10 μL, Hg 2+ Samples of the test solutions with different concentrations (referred to as Sample 1, Sample 2, and Sample 3, respectively) were mixed at room temperature so that the concentration of e-T-rich reached 2 μM. The mixture was left at room temperature for 20 minutes. Then 20 units of Exo III were added to 50 μL and left at 45°C for 20 minutes. It was then heated to 80°C for 10 minutes to inactivate the Exo III. Finally, cool slowly to room temperature (approximately 20 min) for PAGE analysis and electrochemical detection on ITO electrode chips. The results of this example are listed in the following table:
[0113] Sample No
Embodiment 2
[0115] Will contain 2 μM e-T-rich probe, 20 units of Exo III and 10 μL containing different concentrations of Hg 2+ The reaction mixtures of the samples to be tested were incubated in 1x NEBuffer1 buffer at 45 °C for 20 min. It was then heated to 80°C for 10 minutes to inactivate the Exo III. Finally, cool slowly to room temperature (approximately 20 min) for PAGE analysis and electrochemical detection on ITO electrode chips. The results of this example are listed in the following table:
[0116] Sample No
Current size
detected Hg 2+ concentration
Sample 1
1.03
0nM
Sample 2
1.96
0.5nM
Sample 3
2.61
1nM
Sample 4
3.33
5nM
Sample 5
4.18
10nM
Sample 6
4.85
20nM
Sample 7
5.77
40nM
Sample 8
6.66
80nM
[0117] in conclusion:
[0118] The above test examples and examples prove that the detection solution of the present invention contains...
PUM
| Property | Measurement | Unit |
|---|---|---|
| electrical resistivity | aaaaa | aaaaa |
| area | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com