RT-PCR kit for identification and diagnosis of porcine epidemic diarrhea virus and application of RT-PCR kit
A porcine epidemic diarrhea and RT-PCR technology, which is applied in the field of porcine epidemic diarrhea virus RT-PCR differential diagnosis kits, can solve the problems of inability to realize the differential diagnosis of natural infection and vaccine immune strains, and achieves high sensitivity and specificity. strong effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Detection of Piglet Diarrhea Sample and Analysis of the ORF3 Gene Sequence of the Epidemic Strain
[0034] (1) Primer design
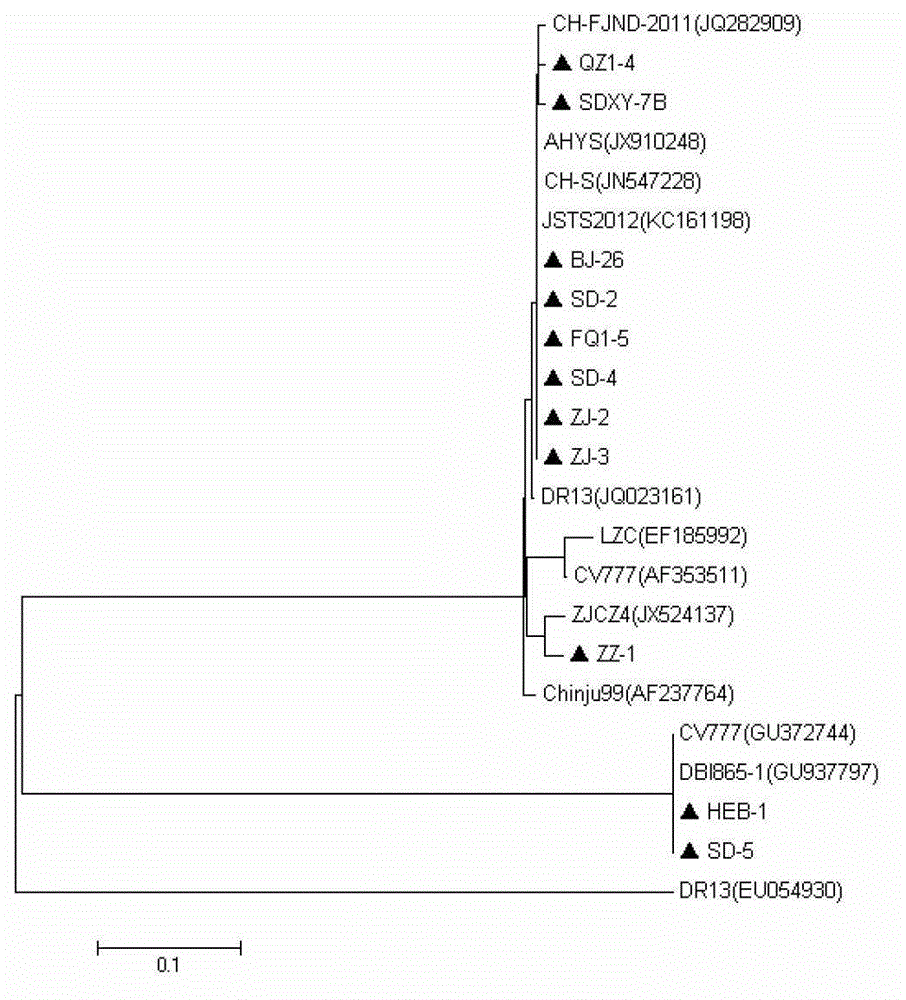
[0035] According to PEDV ZJCZ-4 (GenBank sequence number: JX524137), a pair of amplifying ORF3 whole gene primers ORF3-P1 / ORF3-P2 was designed and synthesized. Referring to the ORF3 gene sequences of PEDV CV777 (GenBank sequence number: AF353511), ZJCZ-4 and vaccine strain DR13 (GenBank sequence number: EU054930), a pair of identification primers ORF3-JD1 / ORF3-JD2 was designed and synthesized, and the identification primers were located at ORF3 The conserved regions at both ends of the deletion region (245nt-294nt) were amplified to cover the entire deletion region. The above two pairs of primers are shown in Table 1, and all primers were synthesized by Shanghai Yingjun Biotechnology Co., Ltd.
[0036] Table 1 Amplification of PEDV ORF3 gene primers and identification primers
[0037]
[0038] (2) RNA extraction and reverse trans...
Embodiment 2
[0046] Optimization of embodiment 2RT-PCR reaction conditions
[0047] (1) Differential diagnosis RT-PCR amplification and determination of optimal Tm value
[0048] The cDNAs of HLJ-1 strain and ZJCZ4 strain were amplified by PCR using identification primers ORF3-JD1 / JD2. The total volume of the PCR reaction is 20 μL, which contains 2 μL of 10×PCR buffer, 1.5 μL of dNTP (2.5 mmol / L), 0.5 μL of upstream and downstream primers, 0.5 μL of rTaq DNA polymerase, and 2 μL of cDNA. Add water to 20 μL. The PCR program was: pre-denaturation at 95°C for 5 min; denaturation at 94°C for 1 min, annealing for 30 s, extension at 72°C for 30 s, and a total of 35 cycles; finally, extension at 72°C for 10 min. The PCR annealing temperature set 5 gradients, namely: 45°C, 48°C, 50°C, 52°C, 55°C. The obtained PCR products were observed by 1% agarose gel electrophoresis.
[0049] (2) Determination of the optimal primer concentration for RT-PCR
[0050] On the basis of the above reaction conditi...
Embodiment 3
[0053] Embodiment 3RT-PCR specific identification test
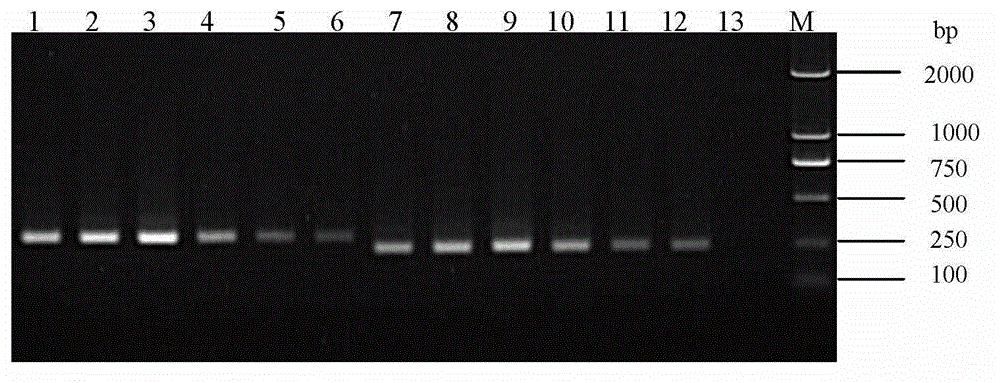
[0054] Extract the RNA of TGEV, PoRV, PRRSV (HuN4 strain), CSFV and PEDV (HLJ-1 strain and ZJCZ4 strain) preserved in our laboratory, and use ORF3-JD1 / JD2 primers to verify the specificity of RT-PCR amplification .
[0055] The results show that only the ZJCZ4 strain and the HLJ-1 strain can amplify specific bands of different sizes, the sizes are about 300bp and 250bp, respectively, and no specific bands occur with other viral nucleic acids as templates (see Figure 5 ). Figure 5 Middle, M: Maker DL2000; 1: TGEV; 2: PoRV; 3: PRRSV (HuN4 strain); 4: CSFV; 5: HLJ-1 strain; 6: ZJCZ4 strain; 7: negative control.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com