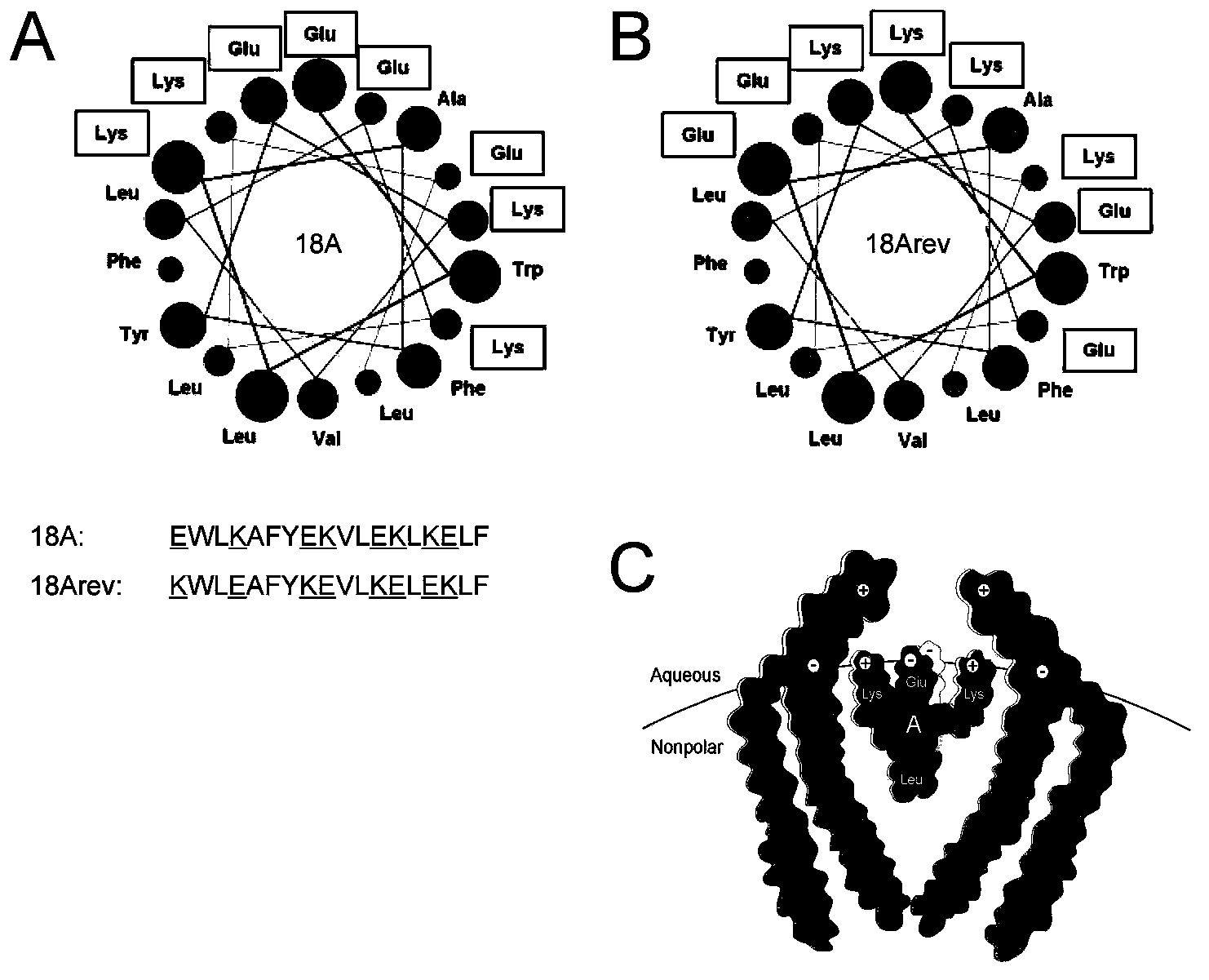
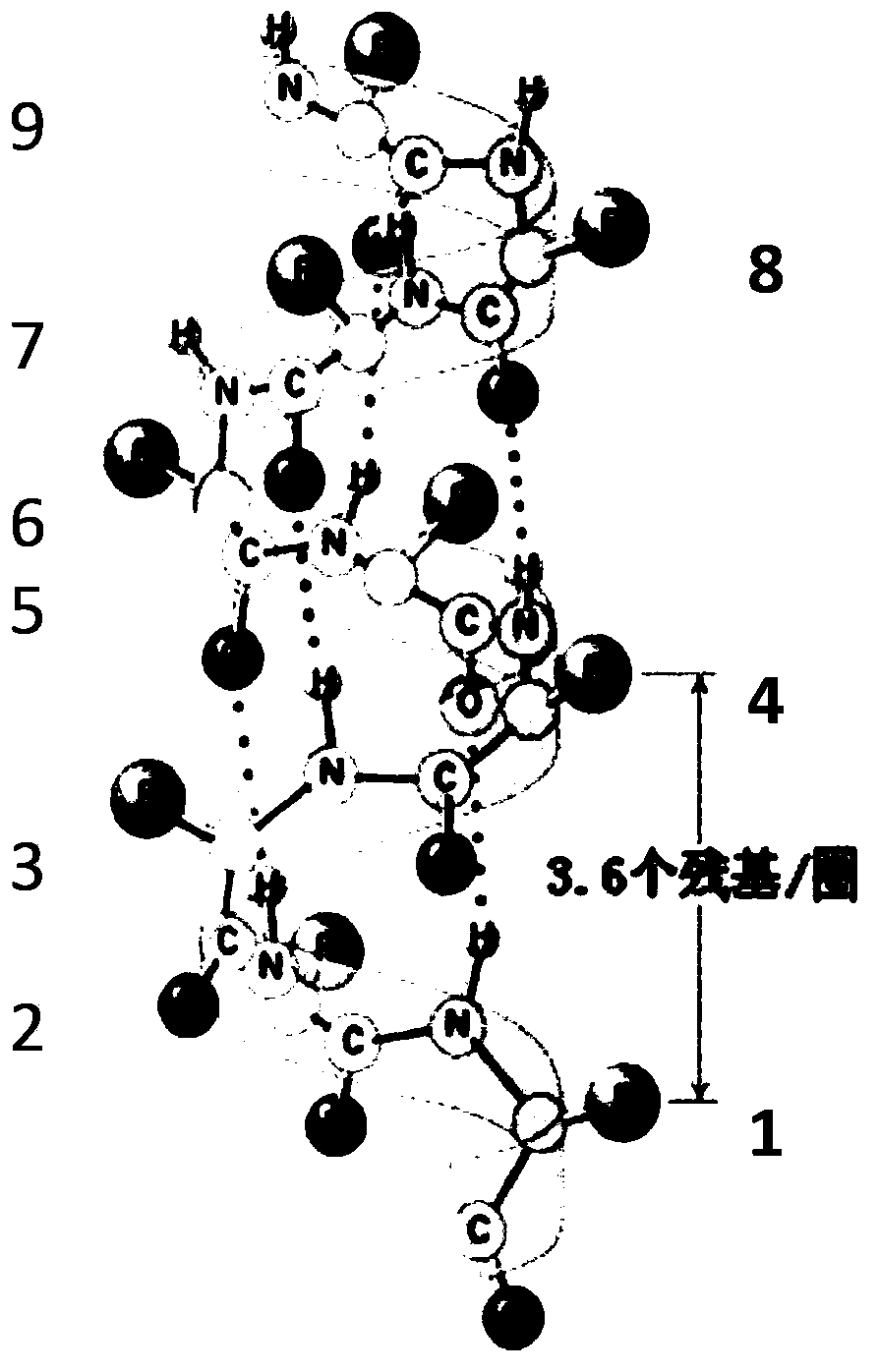
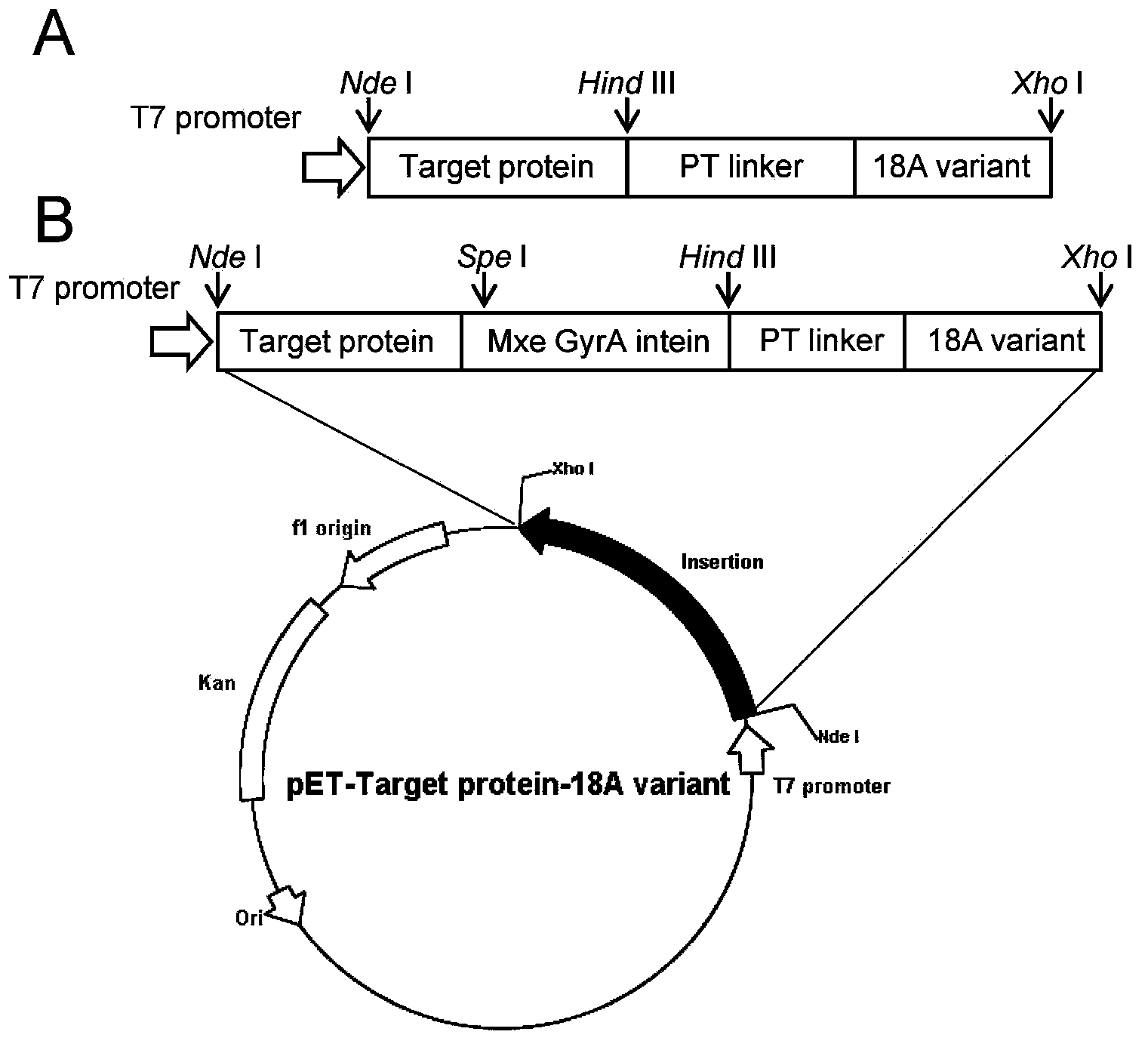
Amphiphilic alpha helix self-assembling peptide and application thereof
A technology selected from, intein, applied in the field of genetic engineering, can solve the problems of restricting the application of inclusion body expression form, complex technology, low yield, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1: Construction of a fusion protein expression vector using Bacillus subtilis lipase A (LipA) and green fluorescent protein (GFP) as target proteins
[0066] 1.1 Amplification of polynucleotide fragments of 18A variants (19 species)
[0067] Firstly, the nucleotide sequence of the PT-type linker and the 18A variant was designed with the online tool DNAworks. The oligonucleotide primers shown in Table 2 were designed by DNAWorks [13] and synthesized, and then the complete sequence encoding the 18A variant (Hind III-linker-18A variant-Xho I) was obtained by overlapping PCR (overlapping PCR) method. polynucleotide sequence.
[0068] Table 2 is used to amplify the primer list of 18A variant
[0069]
[0070]
[0071]
[0072] a The underlined parts of the primers are the recognition sites of restriction endonucleases Hind III and Xho I, respectively.
[0073] Taking one of the variants, 18Av1, as an example, the specific method for amplifying the polynu...
Embodiment 2
[0093] Example 2: Expression, cell growth status and enzyme activity assay of a fusion protein with Bacillus subtilis lipase A (LipA) as the target protein
[0094] 2.1 Induced expression of fusion protein
[0095] The strain constructed in Example 1 (containing plasmids pET-30a(+)-LipA-native, pET-30a(+)-GFP-18A and pET-30a(+)-GFP-18A variants) was inoculated into the 50 μg / mL kanamycin in LB liquid medium, and cultured in a shaker at 37°C to logarithmic phase (OD 600 =0.4-0.6), add 0.2mM IPTG, induce at 30℃ for 6 hours, harvest the cells, and measure the bacterial concentration OD 600 (The following will be 1mL of OD 600 The amount of cells equal to 1 is called 1OD).
[0096] 2.2 Cell growth status
[0097] The growth state of the cells expressing the LipA-18A variant fusion protein is shown in Table 7 and Figure 4 As shown in A.
[0098] Table 7 expresses the OD of LipA-18A and LipA-18A variant fusion protein cells 600 value
[0099] 18A variant LipA-18...
Embodiment 3
[0107] Example 3: Expression of fusion protein with green fluorescent protein (GFP) as the target protein and its intracellular distribution
[0108] The strain constructed in Example 1 was inoculated into LB liquid medium containing 50 μg / mL kanamycin, and cultivated in a shaker at 37°C to logarithmic phase (OD 600 =0.4-0.6), add 0.2mM IPTG, induce at 23°C for 22 hours, and harvest the cells.
[0109] The harvested cells were treated with 4% paraformaldehyde at 4°C for 1 h. Fluorescent confocal microscopy of GFP cells was performed on a Zeiss710 inverted confocal microscope (Zeiss LSM710confocal microscope) with an excitation wavelength of 488nm.
[0110] The intracellular distribution of active enzyme aggregates induced by 18A and 18A variants was as follows Figure 5 shown. It can be clearly seen from the fluorescent photos that the fluorescence of cells expressing GFP-18A fusion protein is mainly distributed on the inner side of the cell membrane. It shows that the GFP...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com