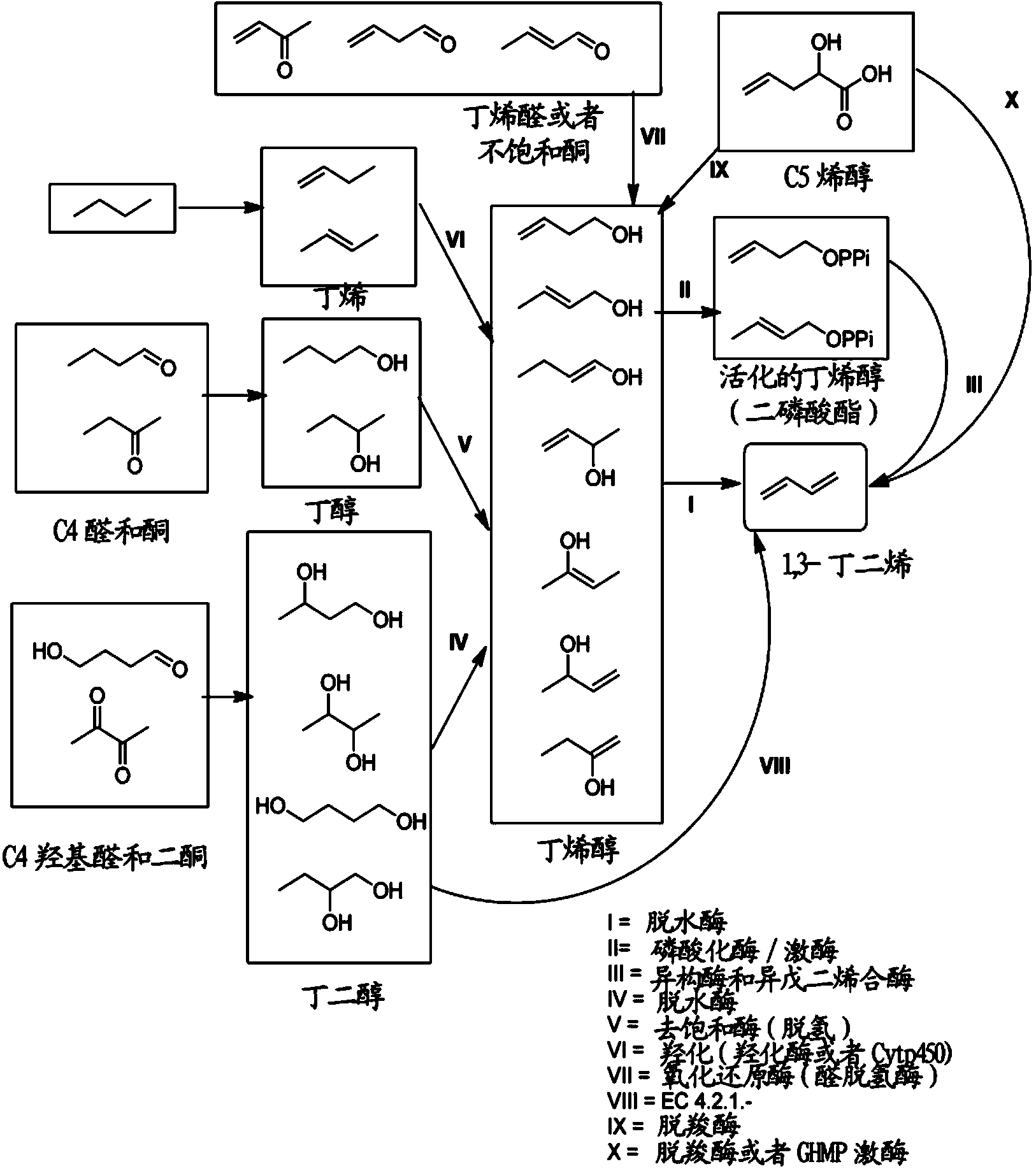
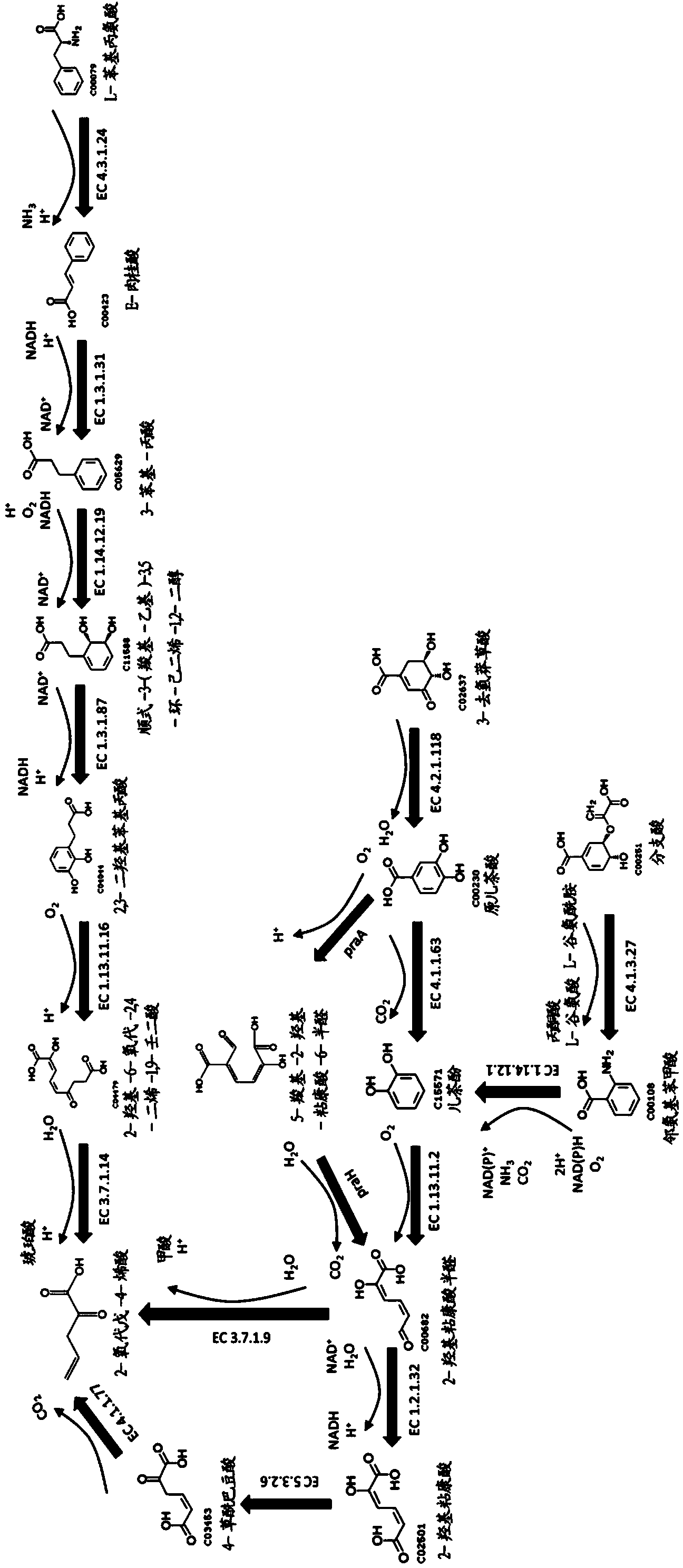
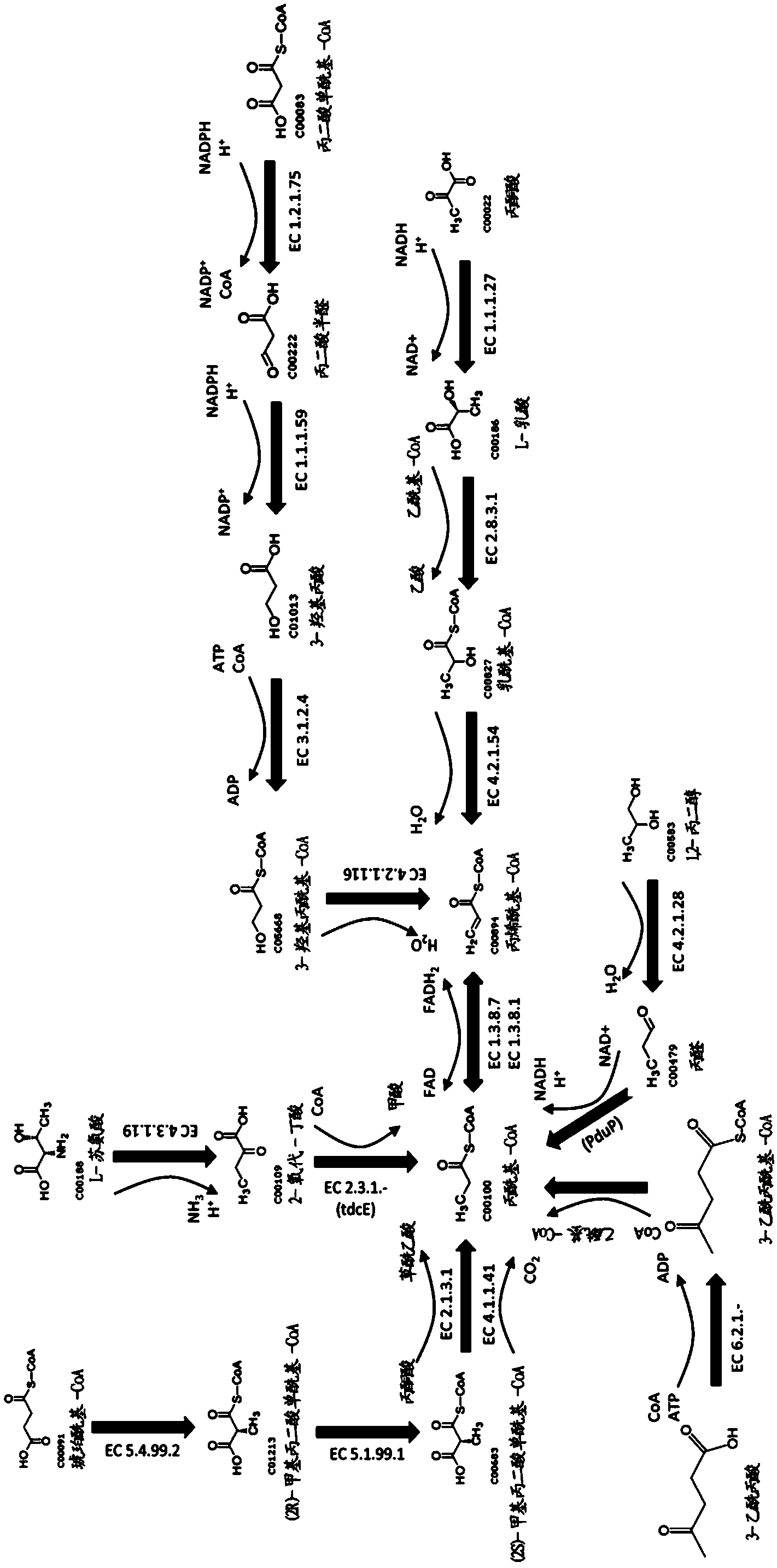
Methods for biosynthesizing 1,3butadiene
A technology of biosynthesis and butadiene, applied in biochemical equipment and methods, waste fuels, enzymes, etc., can solve the problem of less catalytic terminal vinyl groups
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0232] Enzyme activity of mevalonate diphosphate decarboxylase accepting 3-hydroxypent-4-enoate as substrate
[0233] His-tagged MDD genes from S. cerevisiae, S. epidermidis and S. pneumoniae were cloned and expressed in E. coli in shake flask cultures containing Luria Broth medium.
[0234] The pellet from each induced shake flask culture was collected by centrifugation, and the pellet was resuspended and dissolved. Cell debris was separated from the supernatant by centrifugation and filtered using a 0.2 μm filter. The MDD enzyme was purified from the supernatant using Ni-affinity chromatography, concentrated and ultrafiltered by using a 10 kDa polyethersulfone membrane into 50 mM tromethamine buffer (pH=7.5), 100 mM NaCl and 5% (v / v) Buffer exchange with glycerol.
[0235] Native enzyme activity in the presence of 50mM Tris-HCl (pH = 7.5), 100mM NaCl, 5% (v / v) glycerol, 10mM MgCl 2 , 15 mM ATP and 5 mM the natural substrate mevalonate diphosphate (from Sigma Aldrich) buff...
Embodiment 2
[0241] Amino acid residues that enhance mevalonate diphosphate decarboxylase activity in accepting 3-hydroxypent-4-enoate as a substrate
[0242] Figure 13 Amino acid sequences of MDD enzymes from S. cerevisiae, S. epidermidis and S. pneumoniae are provided, with conserved residues within the enzyme's catalytic cleft in bold.
[0243] Using total protein concentration and purity from densitometry, the enzyme concentration was 385 μg / mL for purified MDD from S. cerevisiae and 88 μg / mL for purified MDD from S. pneumoniae.
[0244] Assuming incomplete conversion of 3-hydroxypent-4-enoic acid as an unnatural substrate, the specific conversion of MDD from S. cerevisiae was 809 [(peak area of m / z 54 ion) / (μg MDD)] and The MDD from S. pneumoniae was 3200 [(peak area of m / z 54 ion) / (μg MDD)]. Thus, the specific conversion rate of MDD from S. pneumoniae was about four times that of MDD from S. cerevisiae. The specific conversion of MDD from S. epidermidis was between that of MD...
Embodiment 3
[0248] Enzymatic activity of isoprene synthase accepting trans-2-butenylpyrophosphate as substrate
[0249] The his-tagged isoprene synthase (ISPS) gene from Populus alba was cloned and expressed in E. coli in shake flask cultures containing Luria Broth medium.
[0250] The pellet from each induced shake flask culture was collected by centrifugation, and the pellet was resuspended and dissolved. Cell debris and supernatant were centrifuged and filtered through a 0.2 μm filter. ISPS enzyme variants were purified from supernatants using Ni-affinity chromatography, concentrated and buffer exchanged into 50 mM tromethamine buffer (pH=7.5), 100 mM NaCl and 5% (v / v ) in glycerol.
[0251] The natural enzyme activity was in the presence of 50mM Tris·HCl (pH=7.5), 100mM NaCl, 5% (v / v) glycerol, 20mM MgCl 2 and 5 mM of the natural substrate dimethylallyl diphosphate (from Sigma-Aldrich) at 30°C. Native activity assays were performed in 2 mL septum-sealed vials, thereby allowing acc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com