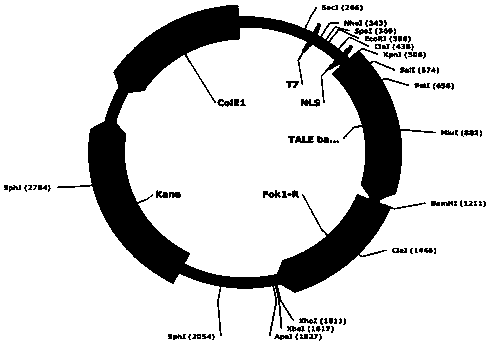
Transcription activator subsample effector nuclease, and coding gene and application thereof
A transcriptional activation and effector technology, applied in the field of genetic engineering, can solve the problems of unexplored therapeutic effect and difficulty in finding TALEN targeting sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Design of TALENs target sequences
[0049] 1. Download human miRNAs: miR-133b, miR-141 and miR-488 gene DNA sequences from miRBase;
[0050] 2. Design TALENs recognition sequence (target sequence):
[0051] According to the sequences obtained from the miRBase database, the recognition sequences of TALENs were determined according to the following principles:
[0052] (1) The 0th base is T (the base before the first in the recognition sequence is the 0th)
[0053] (2) The last base can be AGCT, preferably T
[0054] (3) The length of the recognition sequence is between 13-19
[0055] (4) The length of the spacer sequence between the two recognition sequences is controlled between 14-21 (12,13 are also available, but the efficiency may be lower)
[0056] The position of the designed target sequence is as follows figure 1 The specific sequence is shown in Table 1.
[0057] Table 1
[0058]
Embodiment 2
[0060] Connection between TALENs recognition modules and construction of recombinant vector
[0061] 1. TALENs identification module
[0062] The four recognition modules NI, NG, HD, and NK that respectively recognize bases A, T, C, and G are shown in Table 2 for their sequences.
[0063] Table 2
[0064]
[0065] 2. Identify connections between modules
[0066] Connection strategy: Take the connection of 19 identification modules as an example to illustrate the connection strategy. Since the last module that can recognize the base T is already on the carrier, it only needs to connect 18 modules, and divide the 18 modules into 9 groups in pairs.
[0067] 3. Construction and screening of recombinant vectors
[0068] (1) Construct the left and right arm expression vectors of the nuclease TALEN according to the kit instructions provided by Shanghai Steincell Biotechnology Co., Ltd. Add the reagents corresponding to each module to the above 9 groups in sequence according ...
Embodiment 3
[0071] Human RWPE-1 cells transfected with plasmid
[0072] 1. Add 100 μL Matrigel to each well of a 6-well plate, shake it back and forth to make it cover the bottom of the entire well, and place it in 5% CO 2 30min in the incubator.
[0073] 2. Aspirate the medium in the T25 flask for culturing RWPE-1 cells, aspirate PBS once, add 1mL of 0.25% trypsin, shake back and forth to make it evenly cover the bottom of the flask, and place in 5% CO 2 5min in the incubator.
[0074] 3. Add 1 mL of 10% DMEM to neutralize the trypsin after the digestion is complete, transfer the digested cells to a 15 mL centrifuge tube, count the cells, and centrifuge at 1200 rpm for 5 min.
[0075] 4. Resuspend the cells with an appropriate amount of 10% DMEM, take 2 million RWPE-1 cells and place them in a 6-well plate that has been covered with Matrigel, and add 2 mL of fresh 10% DMEM.
[0076] 5. Subculture and transfect at the same time.
[0077] 6. The constructed miR-133b-TALEN-L and miR-133...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com