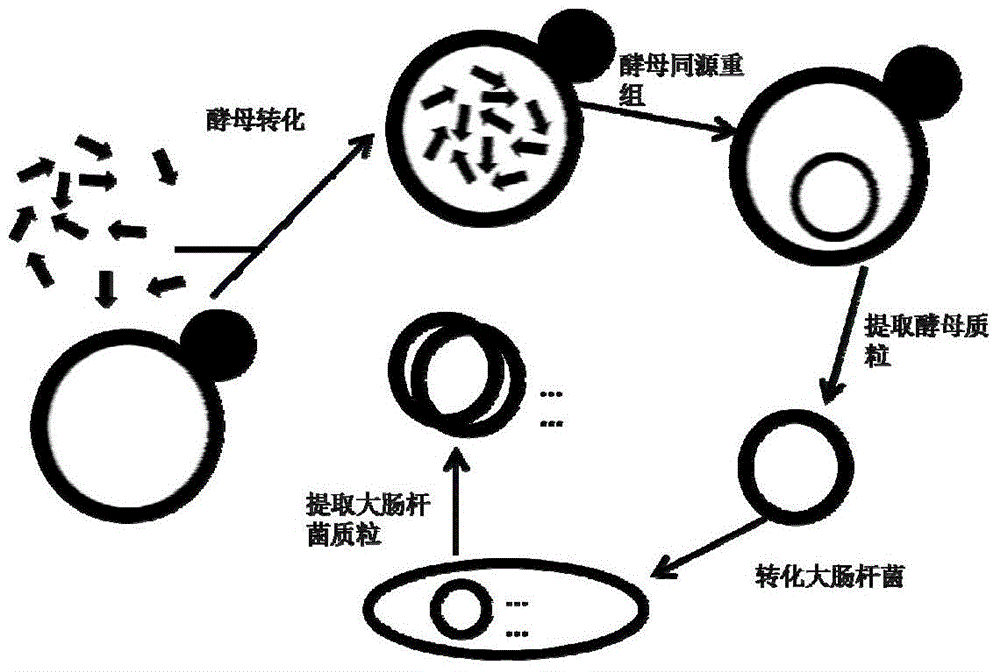
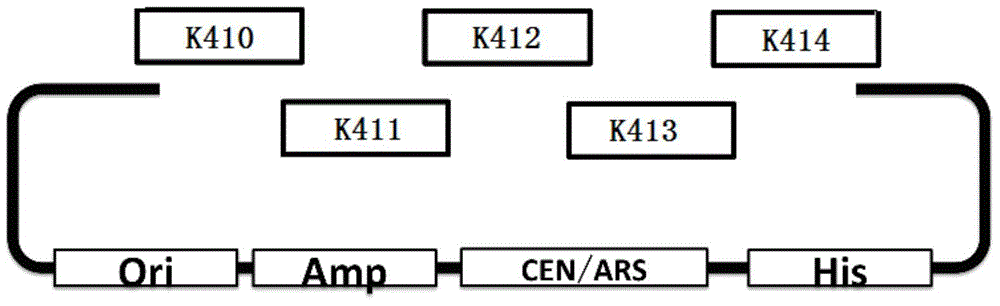
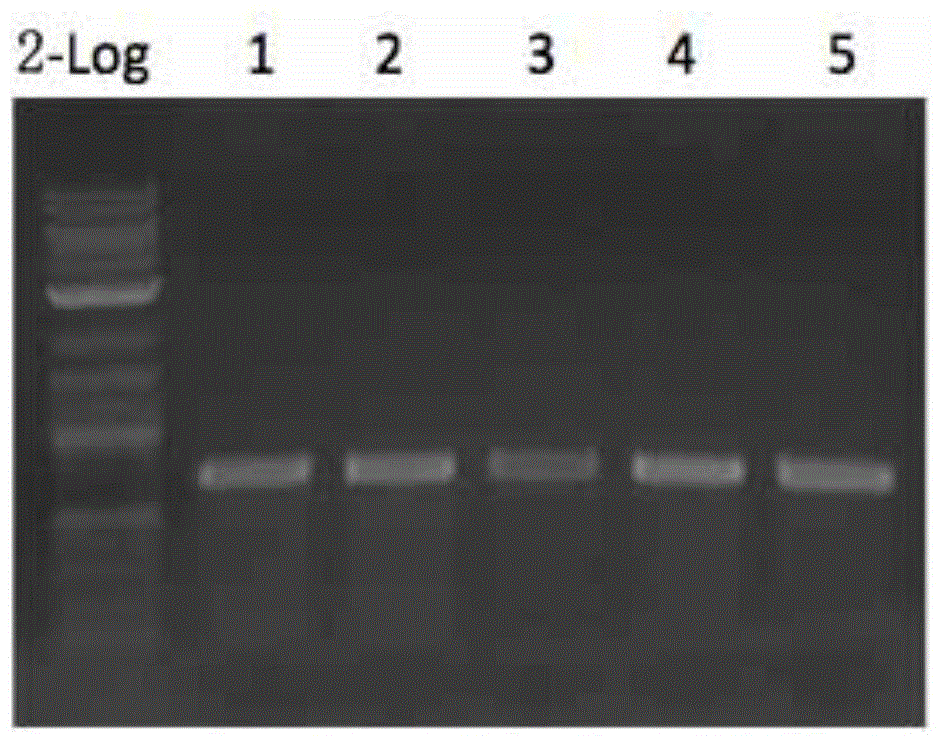
Rapid assembling method of multi-fragment DNA yeast
An assembly method and multi-fragment technology, applied in the field of synthetic biology, can solve the problems of decreased fidelity, increased operational complexity and workload, and prolonged overall time, achieving the effects of low cost, wide application and high success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1: a kind of yeast quick assembly method of multi-fragment DNA, comprises the following steps:
[0045] (1) Co-transform yeast with multiple DNA molecules containing homology arms and linearized yeast shuttle vector:
[0046]① Use K410F shown in SEQ ID NO.1 as the upstream primer, K410R shown in SEQ ID NO.2 as the downstream primer, and pEasy‐K410 shown in SEQ ID NO.3 as the template, using Phusion high-fidelity DNA polymerase Perform PCR to obtain DNA molecular fragment K410; K411F shown in SEQ ID NO.4 is used as an upstream primer, K411R shown in SEQ ID NO.5 is used as a downstream primer, and pEasy‐K411 shown in SEQ ID NO.6 is used as a template, Use Phusion high-fidelity DNA polymerase to perform PCR to obtain DNA molecular fragment K411; use K412F shown in SEQ ID NO.7 as the upstream primer, and K412R shown in SEQ ID NO.8 as the downstream primer, shown in SEQ ID NO.9 The pEasy-K412 of the Phusion high-fidelity DNA polymerase was used as a template to o...
Embodiment 2
[0067] Embodiment 2: a yeast rapid assembly method of multi-fragment DNA, comprising the following steps:
[0068] (1) Co-transform yeast with multiple DNA molecules containing homology arms and linearized yeast shuttle vector:
[0069] ① Use tetR-F shown in SEQ ID NO.19 as the upstream primer, tet-R shown in SEQ ID NO.20 as the downstream primer, and ptetR shown in SEQ ID NO.21 as the template, using Phusion high-fidelity DNA The polymerase performs PCR to obtain the DNA molecular fragment TetR; VioAF shown in SEQ ID NO.22 is used as an upstream primer, VioAR shown in SEQ ID NO.23 is used as a downstream primer, and pVioA shown in SEQ ID NO.24 is used as a template, Use Phusion high-fidelity DNA polymerase to perform PCR to obtain the DNA molecular fragment VioA; VioBF shown in SEQ ID NO.25 is used as the upstream primer, and VioBR shown in SEQ ID NO.26 is used as the downstream primer, shown in SEQ ID NO.27 pVioB as a template, and use Phusion high-fidelity DNA polymerase t...
Embodiment 3
[0082] Embodiment 3: a kind of yeast quick assembly method of multi-fragment DNA, comprises the following steps:
[0083] (1) Co-transform yeast with multiple DNA molecules containing homology arms and linearized yeast shuttle vector:
[0084] ①Using TEF1p-F shown in SEQ ID NO.38 as the upstream primer, TEF1p-R shown in SEQ ID NO.39 as the downstream primer, using the yeast genome as a template, and using Phusion high-fidelity DNA polymerase for PCR to obtain SEQ ID NO. The DNA molecule fragment TEF1p shown in ID NO.56; with PDX1t-F shown in SEQ ID NO.40 as the upstream primer, TDH3p-R shown in SEQ ID NO.41 as the downstream primer, using the yeast genome as a template, using Perform PCR with Phusion high-fidelity DNA polymerase to obtain the DNA fragment PDX1t-TDH3p shown in SEQ ID NO.57; use MPE1t-F shown in SEQ ID NO.42 as the upstream primer, and FBA1p shown in SEQ ID NO.43 ‐R is the downstream primer, using the yeast genome as a template, using Phusion high-fidelity DNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com