Identification method for Klebsiella pneumoniae and adopted primers
A Klebsiella, identification method technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganism measurement/inspection, etc., can solve the problems of complex quantitative measurement instruments, lack of laboratory equipment, high cost, etc. Achieve the effect of simple test steps, improve detection efficiency and save time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Artificially synthesized primer pair K1, K2;
[0039] K1:
[0040] K1-F is: 5-CGATGCTACTTATCCCGACA-3, as shown in SEQ ID NO.1,
[0041] K1-R is: 5-ACCACCAGCAGACGAACTT-3, as shown in SEQ ID NO.2;
[0042] K2:
[0043] K2-F is: 5-CGGAGCGTTTTTCAATCGG-3, as shown in SEQ ID NO.3,
[0044] K2-R is: 5-TGAGCGGGTAATAAATGCGG-3, as shown in SEQ ID NO.4.
Embodiment 2
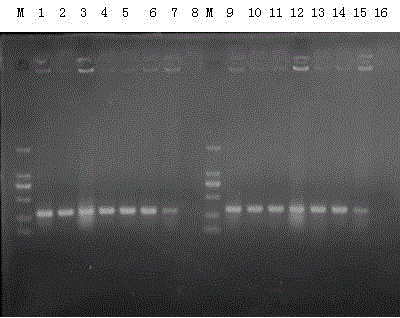
[0046] Recover Klebsiella pneumoniae on ordinary nutrient agar, pick a single colony, shake the bacteria overnight, and use the bacterial solution as a template for PCR reaction. PCR reaction system: 2×Taq PCR MasterMix 25 μL, K1-F and K1-R (upper and lower primers) 2 μL each (or K2-F and K2-R 2 μL each), use bacterial liquid as template 4 μL, and finally use sterilized double steamed Water was added to 50 μL. The reaction conditions are: first step, pre-denaturation at 94°C for 5 minutes; second step, denaturation at 94°C for 30 seconds; third step, annealing at 58°C for 30 seconds; fourth step , extended at a temperature of 72° C. for 1 min; from the second step to the fourth step, a total of 35 cycles were performed; the fifth step: a final extension at a temperature of 72° C. for 7 min; the PCR amplification ended. The amplified product was sequenced, and its sequence was shown in SEQ ID NO.5, which was the specific gene sequence of Klebsiella pneumoniae. It can be...
Embodiment 3
[0051] Specificity test for Escherichia coli
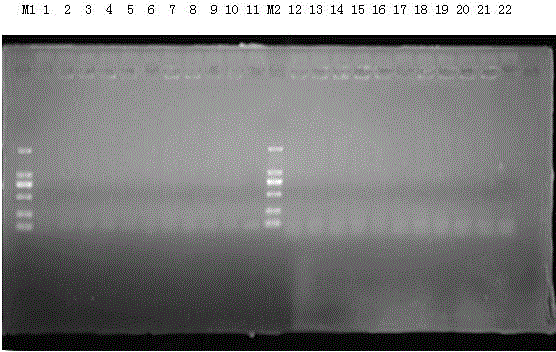
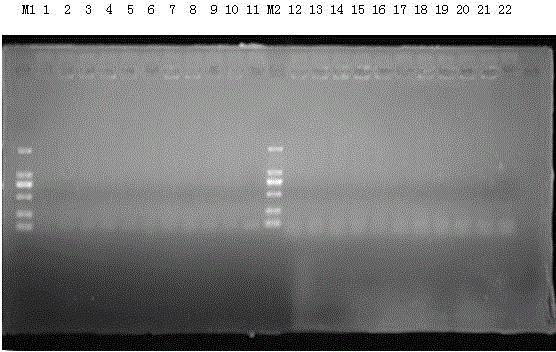
[0052] The 10 strains of Escherichia coli stored in our laboratory were subjected to PCR amplification reaction using the method in Example 2 (the template for PCR is a single colony). After the PCR amplification reaction, 5ul PCR amplification products were taken and subjected to 1.5% (W / V) agarose gel electrophoresis, and the electrophoresis results were photographed on the BIO-RAD ultraviolet gel imaging system; the results are as follows: figure 2 As shown, no band appeared at 303bp. It can be seen that this primer cannot detect Escherichia coli.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com