Quantitative detection method of live bacteria based on FISH technique
A quantitative detection method and technology for live bacteria, applied in the field of microbial detection, can solve the problems of heavy workload, inability to use culture method detection, long time consumption, etc., and achieve the effect of strong universality
Active Publication Date: 2015-04-29
FUZHOU UNIV
View PDF0 Cites 6 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0002] The culture method is currently the most widely used specific detection method for active microorganisms, but the culture method has obvious disadvantages: on the one hand, the method takes a long time, the workload is heavy, and the detection results have a large lag; on the other hand, for feces Bacteria that are not cultivable or in a viable non-culturable state (Viable but Non-culturable, VBNC) in the sample cannot be detected by culture methods
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
[0030] The present invention will be further described below in conjunction with specific examples, but the present invention is not limited thereto.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
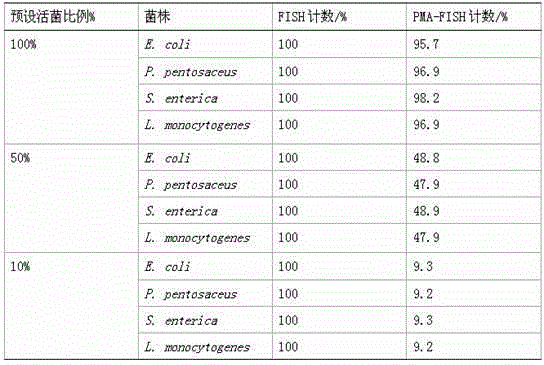
The invention belongs to the field of microbiological detection, and relates to a quantitative detection method of live bacteria based on an FISH technique. The method comprises the following steps: adding a PMA nucleic acid dye for pre-treating before an implementation process of a conventional fluorescence in-situ hybridization method; carrying out covalent binding of nucleic acid in dead bacterium cells and PMA in the PMA treatment process, and sending out orange fluorescence under blue light excitation of a fluorescence microscope, so as to know that the bacteria in the orange fluorescence are dead bacteria in an original system; combining rRNA in live bacterium cells with a fluorescence probe in a hybridization step, and sending out green fluorescence under blue light excitation of the fluorescence microscope by the fluorescence probe, so as to know that the bacteria in the green fluorescence are the live bacteria in the original system. Through improvement of a fluorescent in-situ hybridization technique, the step of adding PMA dye for treating is increased, and effective distinguishing of the dead bacteria and the live bacteria in a sample system is achieved; and a set of method capable of rapidly and quantitatively detecting the live bacteria in the sample is built. The method has relatively high universality in bacterial quantization.
Description
technical field [0001] The invention belongs to the field of microorganism detection and relates to a method for quantitative detection of living bacteria based on FISH technology. Background technique [0002] The culture method is currently the most widely used specific detection method for active microorganisms, but the culture method has obvious disadvantages: on the one hand, the method takes a long time, the workload is heavy, and the detection results have a large lag; on the other hand, for feces Bacteria that are not culturable or in a viable non-culturable state (Viable but Non-culturable, VBNC) in the sample cannot be detected by culture methods. [0003] Fluorescence in situ hybridization (FISH) technology uses fluorescently labeled specific nucleic acid probes to hybridize with corresponding target DNA molecules or RNA molecules in cells, and observes fluorescent signals under a fluorescent microscope or confocal laser scanner to determine the specific probes th...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): C12Q1/68C12Q1/06
CPCC12Q1/6841C12Q1/689C12Q2563/107C12Q2543/10
Inventor 倪莉陈智超刘志彬张雯
Owner FUZHOU UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com