DNA (deoxyribonucleic acid) fragments of ginseng specificity RAPD (random amplified polymorphic DNA) mark, and primer pair and method for identifying SCAR mark of ginseng
A technology of specificity and primer pairs, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as low annealing temperature, discontinuity, and poor repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
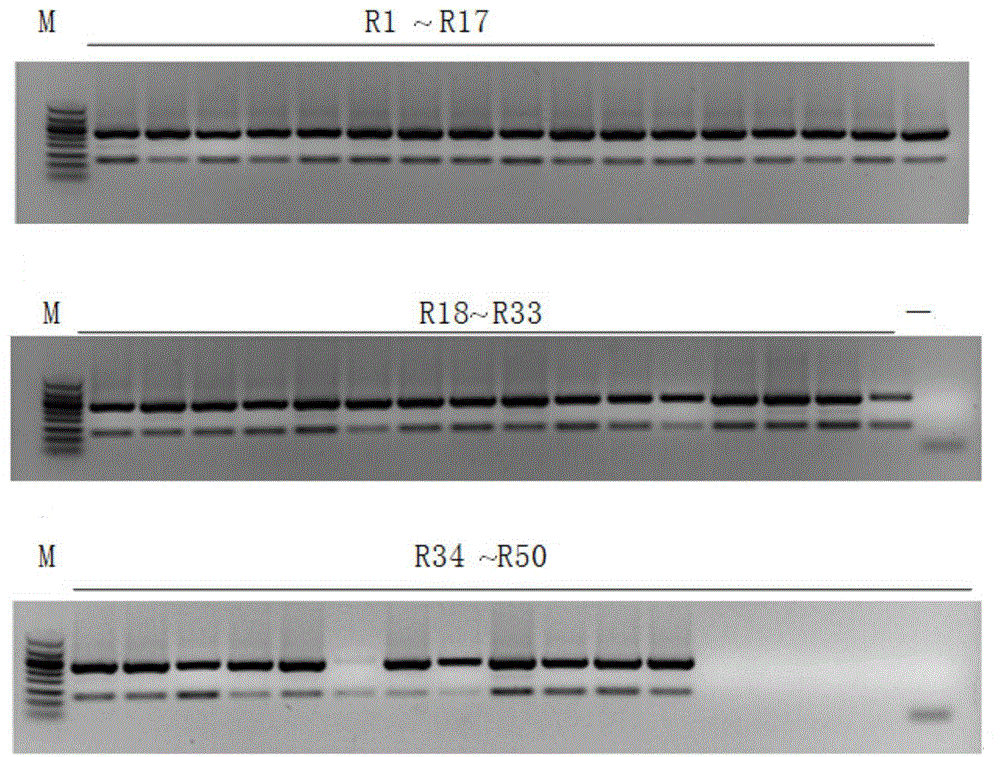
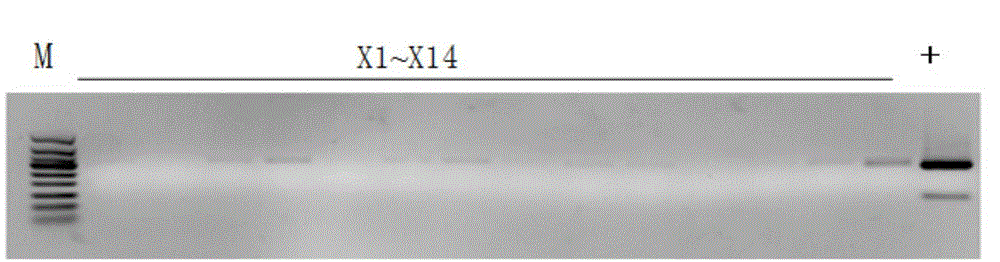
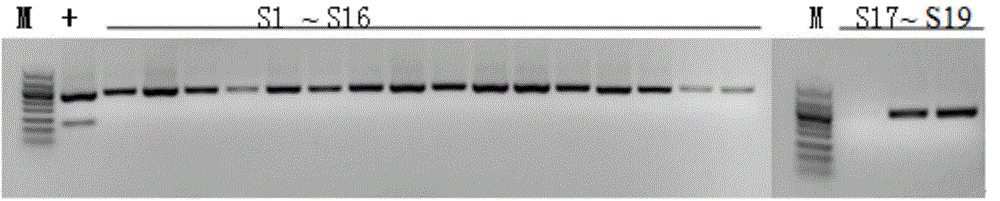
[0032] Materials selected for the experiment of the present invention: all samples in total: 150 batches, of which: 50 batches of ginseng medicinal materials (illustrating that R1~R31 is ginseng, and R32~R50 is ginseng processed with sugar, which is commonly known as red ginseng or white ginseng) ; 14 batches of American ginseng; 19 batches of Panax notoginseng; 9 batches of counterfeit products; 58 batches of other Chinese herbal medicines. (Materials are shown in Table 1 to Table 5)
[0033] In the present invention, 29 samples of ginseng, American ginseng and Panax notoginseng are firstly amplified by PCR using RAPD molecular marker technology, and the specific fragments are obtained by screening. The specific fragments are converted into SCAR markers after recovery, cloning and sequencing, and primer design. .
[0034] The steps of the molecular marker method for identification of ginseng are as follows:
[0035] (1) Extraction of genomic DNA: Wipe and disinfect the surf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com